5LST
 
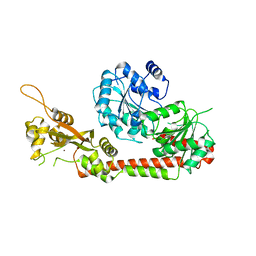 | | Crystal structure of the human RecQL4 helicase. | | Descriptor: | ATP-dependent DNA helicase Q4, ZINC ION | | Authors: | Kaiser, S, Sauer, F, Kisker, C. | | Deposit date: | 2016-09-05 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The structural and functional characterization of human RecQ4 reveals insights into its helicase mechanism.
Nat Commun, 8, 2017
|
|
8BW1
 
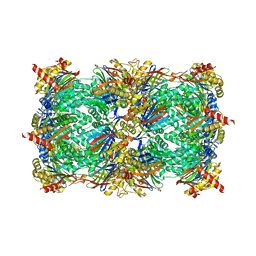 | | Yeast 20S proteasome in complex with an engineered fellutamide derivative (C14QAL) | | Descriptor: | 3-PYRIDIN-4-YL-2,4-DIHYDRO-INDENO[1,2-.C.]PYRAZOLE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Bozhueyuek, K.A.J, Praeve, L, Kegler, C, Kaiser, S, Shi, Y, Kuttenlochner, W, Schenk, L, Groll, M, Hochberg, G.K.A, Bode, H.B. | | Deposit date: | 2022-12-06 | | Release date: | 2023-12-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Evolution-inspired engineering of nonribosomal peptide synthetases.
Science, 383, 2024
|
|
8OWX
 
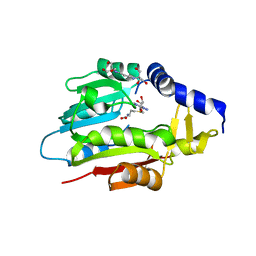 | | Crystal Structure of METTL6 bound to SAH | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, S-ADENOSYL-L-HOMOCYSTEINE, tRNA N(3)-methylcytidine methyltransferase METTL6 | | Authors: | Throll, P, Basu, S, Dolce, L.G, Kowalinski, E. | | Deposit date: | 2023-04-28 | | Release date: | 2024-05-08 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structural basis of tRNA recognition by the m 3 C RNA methyltransferase METTL6 in complex with SerRS seryl-tRNA synthetase.
Nat.Struct.Mol.Biol., 2024
|
|
8OWY
 
 | |
8P7D
 
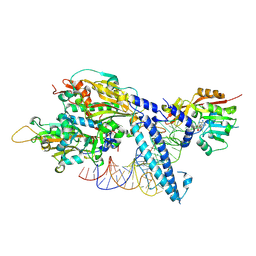 | |
8P7B
 
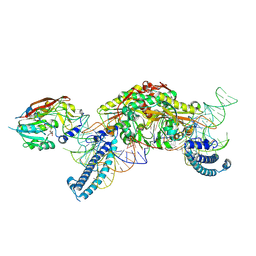 | |
8P7C
 
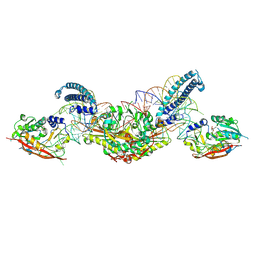 | |
7K6O
 
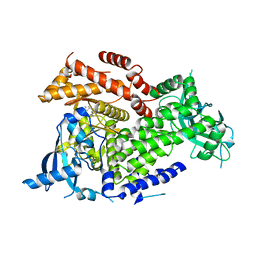 | | Crystal structure of PI3Kalpha inhibitor 10-5429 | | Descriptor: | (3S)-3-[4-(2-aminopyrimidin-5-yl)-2-(morpholin-4-yl)-5,6-dihydro-7H-pyrrolo[2,3-d]pyrimidin-7-yl]-N-methylpyrrolidine-1-sulfonamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Chen, P, Brooun, A, Deng, Y.L, Grodsky, N, Kaiser, S.E. | | Deposit date: | 2020-09-21 | | Release date: | 2021-01-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.738 Å) | | Cite: | Structure-Based Drug Design and Synthesis of PI3K alpha-Selective Inhibitor (PF-06843195).
J.Med.Chem., 64, 2021
|
|
7K71
 
 | | Crystal structure of PI3Kalpha inhibitor 4-0686 | | Descriptor: | 2-(morpholin-4-yl)[4,5'-bipyrimidin]-2'-amine, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Chen, P, Brooun, A, Deng, Y.L, Grodsky, N, Kaiser, S.E. | | Deposit date: | 2020-09-21 | | Release date: | 2021-01-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-Based Drug Design and Synthesis of PI3K alpha-Selective Inhibitor (PF-06843195).
J.Med.Chem., 64, 2021
|
|
7K6N
 
 | | Crystal structure of PI3Kalpha selective Inhibitor 11-1575 | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, tert-butyl (3S)-3-[4-(2-aminopyrimidin-5-yl)-2-(morpholin-4-yl)-5,6-dihydro-7H-pyrrolo[2,3-d]pyrimidin-7-yl]-3-methylpyrrolidine-1-carboxylate | | Authors: | Chen, P, Brooun, A, Deng, Y.L, Grodsky, N, Kaiser, S.E. | | Deposit date: | 2020-09-21 | | Release date: | 2021-01-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structure-Based Drug Design and Synthesis of PI3K alpha-Selective Inhibitor (PF-06843195).
J.Med.Chem., 64, 2021
|
|
7K6M
 
 | | Crystal structure of PI3Kalpha selective Inhibitor PF-06843195 | | Descriptor: | 2,2-difluoroethyl (3S)-3-{[2'-amino-5-fluoro-2-(morpholin-4-yl)[4,5'-bipyrimidin]-6-yl]amino}-3-(hydroxymethyl)pyrrolidine-1-carboxylate, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Chen, P, Brooun, A, Deng, Y.L, Grodsky, N, Kaiser, S.E. | | Deposit date: | 2020-09-21 | | Release date: | 2021-01-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.413 Å) | | Cite: | Structure-Based Drug Design and Synthesis of PI3K alpha-Selective Inhibitor (PF-06843195).
J.Med.Chem., 64, 2021
|
|
2MPC
 
 | |
7KKM
 
 | | Structure of the catalytic domain of tankyrase 1 | | Descriptor: | Poly [ADP-ribose] polymerase, ZINC ION | | Authors: | Gajiwala, K.S, Ryan, K. | | Deposit date: | 2020-10-27 | | Release date: | 2021-01-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Dissecting the molecular determinants of clinical PARP1 inhibitor selectivity for tankyrase1.
J.Biol.Chem., 296, 2021
|
|
7KKN
 
 | | Structure of the catalytic domain of tankyrase 1 in complex with talazoparib | | Descriptor: | (8S,9R)-5-fluoro-8-(4-fluorophenyl)-9-(1-methyl-1H-1,2,4-triazol-5-yl)-2,7,8,9-tetrahydro-3H-pyrido[4,3,2-de]phthalazin-3-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Poly [ADP-ribose] polymerase, ... | | Authors: | Gajiwala, K.S, Ryan, K. | | Deposit date: | 2020-10-27 | | Release date: | 2021-01-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Dissecting the molecular determinants of clinical PARP1 inhibitor selectivity for tankyrase1.
J.Biol.Chem., 296, 2021
|
|
7KK4
 
 | | Structure of the catalytic domain of PARP1 in complex with olaparib | | Descriptor: | 4-(3-{[4-(cyclopropylcarbonyl)piperazin-1-yl]carbonyl}-4-fluorobenzyl)phthalazin-1(2H)-one, Poly [ADP-ribose] polymerase 1 | | Authors: | Gajiwala, K.S, Ryan, K. | | Deposit date: | 2020-10-27 | | Release date: | 2021-01-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Dissecting the molecular determinants of clinical PARP1 inhibitor selectivity for tankyrase1.
J.Biol.Chem., 296, 2021
|
|
7KK3
 
 | | Structure of the catalytic domain of PARP1 in complex with talazoparib | | Descriptor: | (8S,9R)-5-fluoro-8-(4-fluorophenyl)-9-(1-methyl-1H-1,2,4-triazol-5-yl)-2,7,8,9-tetrahydro-3H-pyrido[4,3,2-de]phthalazin-3-one, Poly [ADP-ribose] polymerase 1 | | Authors: | Gajiwala, K.S, Ryan, K. | | Deposit date: | 2020-10-27 | | Release date: | 2021-01-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Dissecting the molecular determinants of clinical PARP1 inhibitor selectivity for tankyrase1.
J.Biol.Chem., 296, 2021
|
|
7Z1K
 
 | | Crystal structure of the SPOC domain of human SHARP (SPEN) in complex with RNA polymerase II CTD heptapeptide phosphorylated on Ser5 | | Descriptor: | Msx2-interacting protein, SER-TYR-SER-PRO-THR-SEP | | Authors: | Appel, L, Grishkovskaya, I, Slade, D, Djinovic-Carugo, K. | | Deposit date: | 2022-02-24 | | Release date: | 2022-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The SPOC domain is a phosphoserine binding module that bridges transcription machinery with co- and post-transcriptional regulators.
Nat Commun, 14, 2023
|
|
7Z27
 
 | |
7KK6
 
 | |
7KKQ
 
 | |
7KK2
 
 | | Structure of the catalytic domain of PARP1 | | Descriptor: | Poly [ADP-ribose] polymerase 1, SULFATE ION | | Authors: | Gajiwala, K.S, Ryan, K. | | Deposit date: | 2020-10-27 | | Release date: | 2021-01-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.695 Å) | | Cite: | Dissecting the molecular determinants of clinical PARP1 inhibitor selectivity for tankyrase1.
J.Biol.Chem., 296, 2021
|
|
7KK5
 
 | |
7KKP
 
 | |
7KKO
 
 | | Structure of the catalytic domain of tankyrase 1 in complex with olaparib | | Descriptor: | 4-(3-{[4-(cyclopropylcarbonyl)piperazin-1-yl]carbonyl}-4-fluorobenzyl)phthalazin-1(2H)-one, Poly [ADP-ribose] polymerase, ZINC ION | | Authors: | Gajiwala, K.S, Ryan, K. | | Deposit date: | 2020-10-27 | | Release date: | 2021-01-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Dissecting the molecular determinants of clinical PARP1 inhibitor selectivity for tankyrase1.
J.Biol.Chem., 296, 2021
|
|
