7KOA
 
 | | Room Temperature Structure of SARS-CoV-2 Nsp10/16 Methyltransferase in a Complex with Cap-0 and SAM Determined by Pink-Beam Serial Crystallography | | Descriptor: | 2'-O-methyltransferase, Non-structural protein 10, P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, ... | | Authors: | Wilamowski, M, Sherrell, D.A, Minasov, G, Shuvalova, L, Lavens, A, Henning, R, Maltseva, N, Rosas-Lemus, M, Kim, Y, Satchell, K.J.F, Srajer, V, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-07 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Room Temperature Structure of SARS-CoV-2 Nsp10/Nsp16 Methyltransferase in a Complex with Cap-0 and SAM Determined by Pink-Beam Serial Crystallography
To Be Published
|
|
1K77
 
 | | Crystal Structure of EC1530, a Putative Oxygenase from Escherichia coli | | Descriptor: | FORMIC ACID, GLYCEROL, Hypothetical protein ygbM, ... | | Authors: | Kim, Y, Skarina, T, Beasley, S, Laskowski, R, Arrowsmith, C.H, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-10-18 | | Release date: | 2002-03-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal structure of Escherichia coli EC1530, a glyoxylate induced protein YgbM.
Proteins, 48, 2002
|
|
1K3R
 
 | | Crystal Structure of the Methyltransferase with a Knot from Methanobacterium thermoautotrophicum | | Descriptor: | conserved protein MT0001 | | Authors: | Zarembinski, T.I, Kim, Y, Peterson, K, Christendat, D, Dharamsi, A, Arrowsmith, C.H, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-10-03 | | Release date: | 2002-05-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Deep trefoil knot implicated in RNA binding found in an archaebacterial protein.
Proteins, 50, 2003
|
|
7L52
 
 | | Crystal Structure of the Metallo Beta Lactamase L1 from Stenotrophomonas maltophilia Determined by Serial Crystallography | | Descriptor: | Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase), ZINC ION | | Authors: | Wilamowski, M, Kim, Y, Sherrell, D.A, Lavens, A, Maltseva, N, Endres, M, Babnigg, G, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-21 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of the Metallo Beta Lactamase L1 from Stenotrophomonas maltophilia Determined by Serial Crystallography
To Be Published
|
|
1KS2
 
 | | Crystal Structure Analysis of the rpiA, Structural Genomics, protein EC1268. | | Descriptor: | protein EC1268, RPIA | | Authors: | Zhang, R, Joachimiak, A, Edwards, A.M, Skarina, T, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-01-10 | | Release date: | 2002-08-14 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of Escherichia coli ribose-5-phosphate isomerase: a ubiquitous enzyme of the pentose phosphate pathway and the Calvin cycle.
STRUCTURE, 11, 2003
|
|
1KJN
 
 | |
1KYT
 
 | | Crystal Structure of Thermoplasma acidophilum 0175 (APC014) | | Descriptor: | CALCIUM ION, hypothetical protein TA0175 | | Authors: | Kim, Y, Joachimiak, A, Edwards, A, Xu, X, Pennycooke, M, Gu, J, Cheung, F, Christendat, D, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-02-05 | | Release date: | 2003-01-21 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Thermoplasma acidophilum 0175 (APC014)
To be published
|
|
1L1S
 
 | | Structure of Protein of Unknown Function MTH1491 from Methanobacterium thermoautotrophicum | | Descriptor: | hypothetical protein MTH1491 | | Authors: | Christendat, D, Saridakis, V, Kim, Y, Kumar, P.A, Xu, X, Semesi, A, Joachimiak, A, Arrowsmith, C.H, Edwards, A.M, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-02-19 | | Release date: | 2002-05-29 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of hypothetical protein MTH1491 from Methanobacterium thermoautotrophicum.
Protein Sci., 11, 2002
|
|
1L3L
 
 | | Crystal structure of a bacterial quorum-sensing transcription factor complexed with pheromone and DNA | | Descriptor: | 3-OXO-OCTANOIC ACID (2-OXO-TETRAHYDRO-FURAN-3-YL)-AMIDE, 5'-D(*GP*AP*TP*GP*TP*GP*CP*AP*GP*AP*TP*CP*TP*GP*CP*AP*CP*AP*TP*C)-3', Transcriptional activator protein traR | | Authors: | Zhang, R, Pappas, T, Brace, J.L, Miller, P.C, Oulmassov, T, Molyneaux, J.M, Anderson, J.C, Bashkin, J.K, Winans, S.C, Joachimiak, A. | | Deposit date: | 2002-02-27 | | Release date: | 2002-07-03 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structure of a bacterial quorum-sensing transcription factor complexed with pheromone and DNA.
Nature, 417, 2002
|
|
1L0Q
 
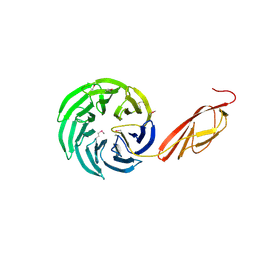 | | Tandem YVTN beta-propeller and PKD domains from an archaeal surface layer protein | | Descriptor: | Surface layer protein | | Authors: | Jing, H, Takagi, J, Liu, J.-H, Lindgren, S, Zhang, R.-G, Joachimiak, A, Wang, J.-H, Springer, T.A. | | Deposit date: | 2002-02-12 | | Release date: | 2002-11-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Archaeal Surface Layer Proteins Contain beta Propeller, PKD, and beta Helix Domains and Are Related to Metazoan Cell Surface Proteins.
Structure, 10, 2002
|
|
7M92
 
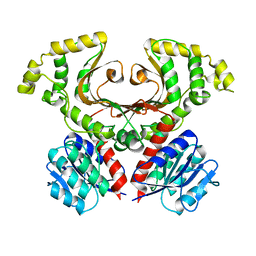 | |
1L8R
 
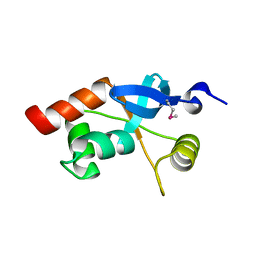 | | Structure of the Retinal Determination Protein Dachshund Reveals a DNA-Binding Motif | | Descriptor: | Dachshund | | Authors: | Kim, S.S, Zhang, R, Braunstein, S.E, Joachimiak, A, Cvekl, A, Hegde, R.S. | | Deposit date: | 2002-03-21 | | Release date: | 2002-06-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of the retinal determination protein Dachshund reveals a DNA binding motif.
Structure, 10, 2002
|
|
1MK4
 
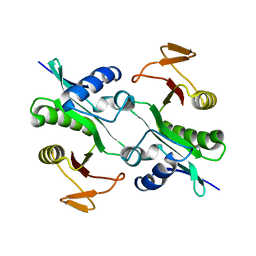 | | Structure of Protein of Unknown Function YqjY from Bacillus subtilis, Probable Acetyltransferase | | Descriptor: | Hypothetical protein yqjY | | Authors: | Zhang, R, Dementiva, I, Mo, A, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-08-28 | | Release date: | 2003-04-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1.7A crystal structure of a hypothetical protein yqjY
from Bacillus subtilis
To be Published
|
|
1MQ8
 
 | | Crystal structure of alphaL I domain in complex with ICAM-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Integrin alpha-L, ... | | Authors: | Shimaoka, M, Xiao, T, Liu, J.-H, Yang, Y, Dong, Y, Jun, C.-D, McCormack, A, Zhang, R, Joachimiak, A, Takagi, J, Wang, J.-H, Springer, T.A. | | Deposit date: | 2002-09-15 | | Release date: | 2003-01-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of the aL I domain and its complex with ICAM-1 reveal a shape-shifting pathway for integrin regulation
Cell(Cambridge,Mass.), 112, 2003
|
|
1MQ9
 
 | | Crystal structure of high affinity alphaL I domain with ligand mimetic crystal contact | | Descriptor: | Integrin alpha-L, MANGANESE (II) ION | | Authors: | Shimaoka, M, Xiao, T, Liu, J.-H, Yang, Y, Dong, Y, Jun, C.-D, McCormack, A, Zhang, R, Joachimiak, A, Takagi, J, Wang, J.-H, Springer, T.A. | | Deposit date: | 2002-09-15 | | Release date: | 2003-01-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the aL I domain and its complex with ICAM-1 reveal a shape-shifting pathway for integrin regulation
Cell(Cambridge,Mass.), 112, 2003
|
|
4PW4
 
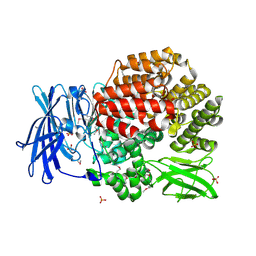 | | Crystal structure of Aminopeptidase N in complex with phosphonic acid analogue of homophenylalanine L-(R)-hPheP | | Descriptor: | Aminopeptidase N, GLYCEROL, IMIDAZOLE, ... | | Authors: | Nocek, B, Mulligan, R, Vassiliou, S, Berlicki, L, Mucha, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-03-18 | | Release date: | 2014-06-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of Aminopeptidase N in complex with phosphonic analogs of homophenylalanine
TO BE PUBLISHED
|
|
4WKY
 
 | | Streptomcyes albus JA3453 oxazolomycin ketosynthase domain OzmN KS2 | | Descriptor: | 1,2-ETHANEDIOL, Beta-ketoacyl synthase, GLYCEROL, ... | | Authors: | Cuff, M.E, Mack, J.C, Endres, M, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-10-03 | | Release date: | 2014-10-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and evolutionary relationships of "AT-less" type I polyketide synthase ketosynthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4Q63
 
 | | Crystal Structure of Legionella Uncharacterized Protein Lpg0364 | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Kim, Y, Evdokimova, E, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-04-21 | | Release date: | 2014-05-07 | | Method: | X-RAY DIFFRACTION (1.953 Å) | | Cite: | Crystal Structure of Legionella Uncharacterized Protein Lpg0364
To be Published
|
|
4X3Z
 
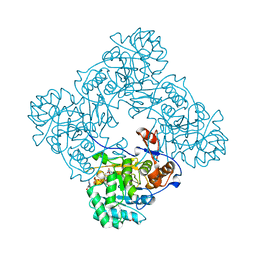 | | Inosine 5'-monophosphate dehydrogenase from Vibrio cholerae, deletion mutant, in complex with XMP and NAD | | Descriptor: | GLYCEROL, Inosine-5'-monophosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Osipiuk, J, MALTSEVA, N, KIM, Y, Mulligan, R, MAKOWSKA-GRZYSKA, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-12-02 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Inosine 5'-monophosphate dehydrogenase from Vibrio cholerae, deletion mutant, in complex with XMP and NAD
to be published
|
|
4QIR
 
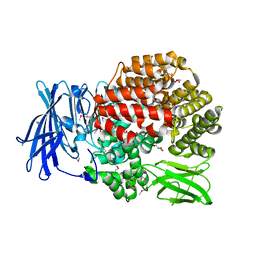 | | Crystal structure of Aminopeptidase N in complex with the phosphinic dipeptide analogue LL-(R,S)-2-(pyridin-3-yl)ethylGlyP[CH2]Phe | | Descriptor: | 3-{[(R)-1-amino-3-(pyridin-3-yl)propyl](hydroxy)phosphoryl}-(S)-2-benzylpropanoic acid, Aminopeptidase N, GLYCEROL, ... | | Authors: | Nocek, B, Joachimiak, A, Berlicki, L, Vassiliou, S, Mucha, A. | | Deposit date: | 2014-06-01 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Structure-guided, single-point modifications in the phosphinic dipeptide structure yield highly potent and selective inhibitors of neutral aminopeptidases.
J.Med.Chem., 57, 2014
|
|
4X2R
 
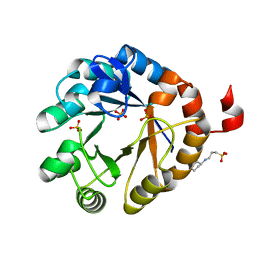 | | Crystal structure of PriA from Actinomyces urogenitalis | | Descriptor: | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, PHOSPHATE ION | | Authors: | MICHALSKA, K, VERDUZCO-CASTRO, E.A, ENDRES, M, BARONA-GOMEZ, F, JOACHIMIAK, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-11-26 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Evolution of substrate specificity in a retained enzyme driven by gene loss.
Elife, 6, 2017
|
|
4QPE
 
 | | Crystal structure of Aminopeptidase N in complex with N-cyclohexyl-1,2-diaminoethylphosphonic acid | | Descriptor: | Aminopeptidase N, SULFATE ION, ZINC ION, ... | | Authors: | Nocek, B, Mulligan, R, Berlicki, L, Vassilious, S, Mucha, A, Joachimiak, A. | | Deposit date: | 2014-06-23 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | Structure-guided, single-point modifications in the phosphinic dipeptide structure yield highly potent and selective inhibitors of neutral aminopeptidases.
J.Med.Chem., 57, 2014
|
|
4QME
 
 | | Crystal structure of Aminopeptidase N in complex with the phosphinic dipeptide analogue LL-(R,S)-hPheP[CH2]Phe | | Descriptor: | (2S)-3-[(S)-[(1R)-1-amino-3-phenylpropyl](hydroxy)phosphoryl]-2-benzylpropanoic acid, Aminopeptidase N, GLYCEROL, ... | | Authors: | Nocek, B, Vassilious, S, Mulligan, R, Berlicki, L, Mucha, A, Joachimiak, A. | | Deposit date: | 2014-06-16 | | Release date: | 2014-10-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Structure-guided, single-point modifications in the phosphinic dipeptide structure yield highly potent and selective inhibitors of neutral aminopeptidases.
J.Med.Chem., 57, 2014
|
|
4QN8
 
 | | The crystal structure of an effector protein VipE from Legionella pneumophila subsp. pneumophila str. Philadelphia 1 | | Descriptor: | BETA-MERCAPTOETHANOL, VipE | | Authors: | Tan, K, Xu, X, Cui, H, Liu, S, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-06-17 | | Release date: | 2014-07-16 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | The crystal structure of an effector protein VipE from Legionella pneumophila subsp. pneumophila str. Philadelphia 1
To be Published
|
|
4X9S
 
 | | CRYSTAL STRUCTURE OF HISAP FROM STREPTOMYCES SP. MG1 | | Descriptor: | Phosphoribosyl isomerase A, SULFATE ION | | Authors: | MICHALSKA, K, VERDUZCO-CASTRO, E.A, ENDRES, M, BARONA-GOMEZ, F, JOACHIMIAK, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-12-11 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Co-occurrence of analogous enzymes determines evolution of a novel ( beta alpha )8-isomerase sub-family after non-conserved mutations in flexible loop.
Biochem. J., 473, 2016
|
|
