6V4R
 
 | |
6OZ9
 
 | | Ebola virus glycoprotein in complex with EBOV-520 Fab | | Descriptor: | EBOV-520 Fab heavy chain, EBOV-520 Fab light chain, Envelope glycoprotein, ... | | Authors: | Milligan, J.C, Altman, P.X, Hui, S, Hastie, K.M, Gilchuk, P, Crowe, J.E, Saphire, E.O. | | Deposit date: | 2019-05-15 | | Release date: | 2020-03-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.462 Å) | | Cite: | Analysis of a Therapeutic Antibody Cocktail Reveals Determinants for Cooperative and Broad Ebolavirus Neutralization.
Immunity, 52, 2020
|
|
6P9J
 
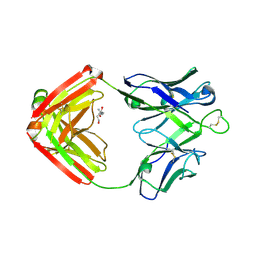 | | crystal structure of human anti staphylococcus aureus antibody STAU-229 Fab | | Descriptor: | TRIS(HYDROXYETHYL)AMINOMETHANE, human anti staphylococcus aureus antibody STAU-229 Fab heavy chain, human anti staphylococcus aureus antibody STAU-229 Fab light chain | | Authors: | Dong, J, Crowe, J.E. | | Deposit date: | 2019-06-10 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Human V H 1-69 Gene-Encoded Human Monoclonal Antibodies against Staphylococcus aureus IsdB Use at Least Three Distinct Modes of Binding To Inhibit Bacterial Growth and Pathogenesis.
Mbio, 10, 2019
|
|
6P9I
 
 | | crystal structure of human anti staphylococcus aureus antibody STAU-399 Fab | | Descriptor: | human anti staphylococcus aureus antibody STAU-399 Fab heavy chain, human anti staphylococcus aureus antibody STAU-399 Fab light chain | | Authors: | Dong, J, Crowe, J.E. | | Deposit date: | 2019-06-10 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Human V H 1-69 Gene-Encoded Human Monoclonal Antibodies against Staphylococcus aureus IsdB Use at Least Three Distinct Modes of Binding To Inhibit Bacterial Growth and Pathogenesis.
Mbio, 10, 2019
|
|
6VY4
 
 | |
6P9H
 
 | |
6V4Q
 
 | | Crystal structure of a MR78-like antibody naive-1 Fab | | Descriptor: | GLYCEROL, ISOPROPYL ALCOHOL, Naive-1 Fab heavy chain, ... | | Authors: | Bozhanova, N.G, Crowe, J.E, Meiler, J. | | Deposit date: | 2019-11-28 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Discovery of Marburg virus neutralizing antibodies from virus-naive human antibody repertoires using large-scale structural predictions.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VY6
 
 | |
6VY5
 
 | |
8OXW
 
 | |
8OXX
 
 | |
8OXV
 
 | |
8OXY
 
 | |
8RIV
 
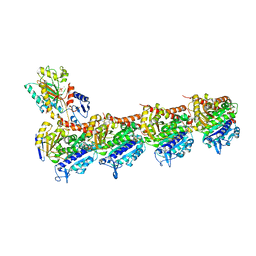 | | T2R-TTL-1-K08 complex | | Descriptor: | (4-fluoranyl-2-methyl-1~{H}-indol-5-yl) 3,4,5-trimethoxybenzenesulfonate, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Boiarska, Z, Homer, J.A, Steinmetz, M.O, Moses, J.E, Prota, A.E.P. | | Deposit date: | 2023-12-19 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Modular synthesis of functional libraries by accelerated SuFEx click chemistry.
Chem Sci, 15, 2024
|
|
8RIW
 
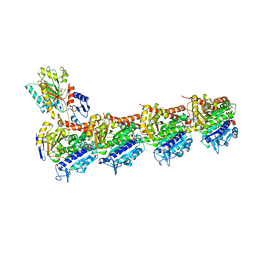 | | T2R-TTL-1-L01 complex | | Descriptor: | (2-methyl-1~{H}-indol-5-yl) 3,4,5-trimethoxybenzenesulfonate, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Prota, A.E.P, Boiarska, Z, Homer, J.A, Steinmetz, M.O, Moses, J.E. | | Deposit date: | 2023-12-19 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Modular synthesis of functional libraries by accelerated SuFEx click chemistry.
Chem Sci, 15, 2024
|
|
8SAG
 
 | |
8SAF
 
 | |
8SZE
 
 | | Crystal structure of Yersinia pestis dihydrofolate reductase in complex with Trimethoprim | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Dihydrofolate reductase, ... | | Authors: | Shaw, G.X, Cherry, S, Tropea, J.E, Waugh, D.S, Ji, X. | | Deposit date: | 2023-05-29 | | Release date: | 2023-06-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Yersinia pestis dihydrofolate reductase in complex with Trimethoprim
To be published
|
|
8SZD
 
 | | Crystal structure of Yersinia pestis dihydrofolate reductase at 1.25-A resolution | | Descriptor: | CHLORIDE ION, Dihydrofolate reductase, MAGNESIUM ION, ... | | Authors: | Shaw, G.X, Cherry, S, Tropea, J.E, Waugh, D.S, Ji, X. | | Deposit date: | 2023-05-29 | | Release date: | 2023-06-07 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structure of Yersinia pestis dihydrofolate reductase at 1.25-A resolution
To be published
|
|
8T0B
 
 | |
8T1N
 
 | | Micro-ED Structure of a Novel Domain of Unknown Function Solved with AlphaFold | | Descriptor: | DUF1842 domain-containing protein | | Authors: | Miller, J.E, Cascio, D, Sawaya, M.R, Cannon, K.A, Rodriguez, J.A, Yeates, T.O. | | Deposit date: | 2023-06-02 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-10 | | Method: | ELECTRON CRYSTALLOGRAPHY (3 Å) | | Cite: | AlphaFold-assisted structure determination of a bacterial protein of unknown function using X-ray and electron crystallography.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8T1M
 
 | | Novel Domain of Unknown Function Solved with AlphaFold | | Descriptor: | DUF1842 domain-containing protein | | Authors: | Miller, J.E, Agdanowski, M.P, Cascio, D, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2023-06-02 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | AlphaFold-assisted structure determination of a bacterial protein of unknown function using X-ray and electron crystallography.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8SD7
 
 | | Carbonic anhydrase II radiation damage RT 61-90 | | Descriptor: | Carbonic anhydrase 2, ZINC ION | | Authors: | Combs, J.E, Mckenna, R. | | Deposit date: | 2023-04-06 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.704 Å) | | Cite: | XFEL structure of carbonic anhydrase II: a comparative study of XFEL, NMR, X-ray and neutron structures.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8SD6
 
 | | Carbonic anhydrase II radiation damage RT 31-60 | | Descriptor: | Carbonic anhydrase 2, ZINC ION | | Authors: | Combs, J.E, Mckenna, R. | | Deposit date: | 2023-04-06 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.397 Å) | | Cite: | XFEL structure of carbonic anhydrase II: a comparative study of XFEL, NMR, X-ray and neutron structures.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8SD9
 
 | | Carbonic anhydrase II radiation damage RT 121-150 | | Descriptor: | Carbonic anhydrase 2, ZINC ION | | Authors: | Combs, J.E, Mckenna, R. | | Deposit date: | 2023-04-06 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | XFEL structure of carbonic anhydrase II: a comparative study of XFEL, NMR, X-ray and neutron structures.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
