5XYF
 
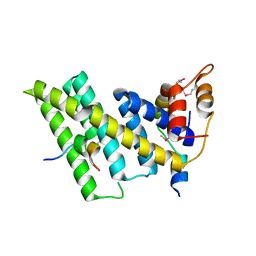 | | Crystal structure of the human TIN2-TPP1-TRF2 telomeric complex | | Descriptor: | Adrenocortical dysplasia protein homolog, TERF1-interacting nuclear factor 2, Telomeric repeat-binding factor 2 | | Authors: | Hu, C, Chen, Y. | | Deposit date: | 2017-07-07 | | Release date: | 2017-12-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Structural and functional analyses of the mammalian TIN2-TPP1-TRF2 telomeric complex.
Cell Res., 27, 2017
|
|
7LFS
 
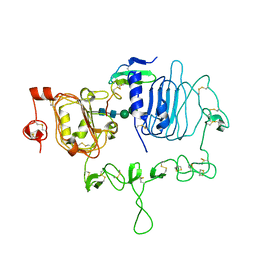 | | Crystal structure of the epidermal growth factor receptor extracellular region with A265V mutation in complex with epiregulin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform 4 of Epidermal growth factor receptor, ... | | Authors: | Hu, C, Leche II, C.A, Stayrook, S.E, Ferguson, K.M, Lemmon, M.A. | | Deposit date: | 2021-01-18 | | Release date: | 2021-11-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Glioblastoma mutations alter EGFR dimer structure to prevent ligand bias.
Nature, 602, 2022
|
|
7LEN
 
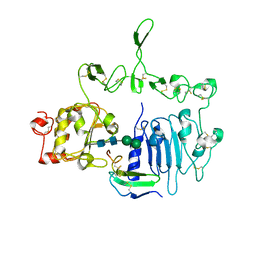 | | Crystal structure of the epidermal growth factor receptor extracellular region with R84K mutation in complex with epiregulin crystallized with trehalose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hu, C, Leche II, C.A, Stayrook, S.E, Ferguson, K.M, Lemmon, M.A. | | Deposit date: | 2021-01-14 | | Release date: | 2021-11-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Glioblastoma mutations alter EGFR dimer structure to prevent ligand bias.
Nature, 602, 2022
|
|
7LFR
 
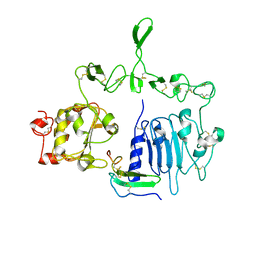 | | Crystal structure of the epidermal growth factor receptor extracellular region with R84K mutation in complex with epiregulin crystallized with spermine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Epidermal growth factor receptor, Proepiregulin, ... | | Authors: | Hu, C, Leche II, C.A, Stayrook, S.E, Ferguson, K.M, Lemmon, M.A. | | Deposit date: | 2021-01-18 | | Release date: | 2021-11-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Glioblastoma mutations alter EGFR dimer structure to prevent ligand bias.
Nature, 602, 2022
|
|
8D8N
 
 | | gRAMP non-match PFS target RNA | | Descriptor: | RAMP superfamily protein, RNA (35-MER), RNA (5'-R(P*UP*CP*CP*GP*GP*GP*GP*CP*AP*GP*AP*AP*AP*AP*UP*UP*GP*GP*AP*CP*A)-3'), ... | | Authors: | Hu, C, Nam, K.H, Schuler, G, Ke, A. | | Deposit date: | 2022-06-08 | | Release date: | 2022-08-31 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Craspase is a CRISPR RNA-guided, RNA-activated protease.
Science, 377, 2022
|
|
8D9H
 
 | | gRAMP-TPR-CHAT match PFS target RNA(Craspase) | | Descriptor: | CHAT domain protein, PHOSPHATE ION, RAMP superfamily protein, ... | | Authors: | Hu, C, Nam, K.H, Schuler, G, Ke, A. | | Deposit date: | 2022-06-09 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Craspase is a CRISPR RNA-guided, RNA-activated protease.
Science, 377, 2022
|
|
8D9E
 
 | | gRAMP-match PFS target | | Descriptor: | RAMP superfamily protein, RNA (36-MER), RNA (5'-R(P*UP*CP*CP*GP*GP*GP*GP*CP*AP*GP*AP*AP*AP*AP*UP*UP*GP*GP*GP*UP*A)-3'), ... | | Authors: | Hu, C, Nam, K.H, Schuler, G, Ke, A. | | Deposit date: | 2022-06-09 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Craspase is a CRISPR RNA-guided, RNA-activated protease.
Science, 377, 2022
|
|
8D97
 
 | | Apo gRAMP | | Descriptor: | RAMP superfamily protein, RNA (42-MER), ZINC ION | | Authors: | Hu, C, Nam, K.H, Schuler, G, Ke, A. | | Deposit date: | 2022-06-09 | | Release date: | 2023-06-14 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Craspase is a CRISPR RNA-guided, RNA-activated protease.
Science, 377, 2022
|
|
8D9F
 
 | | gRAMP-TPR-CHAT (Craspase) | | Descriptor: | CHAT domain protein, RAMP superfamily protein, RNA (33-MER), ... | | Authors: | Hu, C, Nam, K.H, Schuler, G, Ke, A. | | Deposit date: | 2022-06-09 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Craspase is a CRISPR RNA-guided, RNA-activated protease.
Science, 377, 2022
|
|
8D9G
 
 | | gRAMP-TPR-CHAT Non match PFS target RNA(Craspase) | | Descriptor: | CHAT domain protein, RAMP superfamily protein, RNA (36-MER), ... | | Authors: | Hu, C, Nam, K.H, Schuler, G, Ke, A. | | Deposit date: | 2022-06-09 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Craspase is a CRISPR RNA-guided, RNA-activated protease.
Science, 377, 2022
|
|
8D9I
 
 | | gRAMP non-matching PFS-with Mg | | Descriptor: | RAMP superfamily protein, RNA (35-MER), RNA (5'-R(P*UP*CP*CP*GP*GP*GP*GP*CP*AP*GP*AP*AP*AP*AP*UP*UP*GP*GP*A)-3'), ... | | Authors: | Hu, C, Nam, K.H, Schuler, G, Ke, A. | | Deposit date: | 2022-06-09 | | Release date: | 2023-06-14 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Craspase is a CRISPR RNA-guided, RNA-activated protease.
Science, 377, 2022
|
|
8G9S
 
 | | Exploiting Activation and Inactivation Mechanisms in Type I-C CRISPR-Cas3 for Genome Editing Applications | | Descriptor: | AcrIC8, Cas11, Cas5, ... | | Authors: | Hu, C, Nam, K.H, Ke, A. | | Deposit date: | 2023-02-22 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Exploiting activation and inactivation mechanisms in type I-C CRISPR-Cas3 for genome-editing applications.
Mol.Cell, 84, 2024
|
|
8G9T
 
 | | Exploiting Activation and Inactivation Mechanisms in Type I-C CRISPR-Cas3 for Genome Editing Applications | | Descriptor: | AcrIC9, Cas11, Cas5, ... | | Authors: | Hu, C, Nam, K.H, Ke, A. | | Deposit date: | 2023-02-22 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Exploiting activation and inactivation mechanisms in type I-C CRISPR-Cas3 for genome-editing applications.
Mol.Cell, 84, 2024
|
|
8G9U
 
 | | Exploiting Activation and Inactivation Mechanisms in Type I-C CRISPR-Cas3 for Genome Editing Applications | | Descriptor: | CRISPR-associated protein, Csd1 family, Csd2 family, ... | | Authors: | Hu, C, Nam, K.H, Ke, A. | | Deposit date: | 2023-02-22 | | Release date: | 2024-03-06 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Exploiting activation and inactivation mechanisms in type I-C CRISPR-Cas3 for genome-editing applications.
Mol.Cell, 84, 2024
|
|
8GAF
 
 | | Exploiting Activation and Inactivation Mechanisms in Type I-C CRISPR-Cas3 for Genome Editing Applications | | Descriptor: | Cas11, Cas5, Cas7, ... | | Authors: | Hu, C, Nam, K.H, Ke, A. | | Deposit date: | 2023-02-22 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Exploiting activation and inactivation mechanisms in type I-C CRISPR-Cas3 for genome-editing applications.
Mol.Cell, 84, 2024
|
|
8GAM
 
 | | Exploiting Activation and Inactivation Mechanisms in Type I-C CRISPR-Cas3 for Genome Editing Applications | | Descriptor: | Cas11, Cas5, Cas7, ... | | Authors: | Hu, C, Nam, K.H, Ke, A. | | Deposit date: | 2023-02-23 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Exploiting activation and inactivation mechanisms in type I-C CRISPR-Cas3 for genome-editing applications.
Mol.Cell, 84, 2024
|
|
8GAN
 
 | | Exploiting Activation and Inactivation Mechanisms in Type I-C CRISPR-Cas3 for Genome Editing Applications | | Descriptor: | Cas11, Cas5, Cas7, ... | | Authors: | Hu, C, Nam, K.H, Ke, A. | | Deposit date: | 2023-02-23 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Exploiting activation and inactivation mechanisms in type I-C CRISPR-Cas3 for genome-editing applications.
Mol.Cell, 84, 2024
|
|
5YBX
 
 | | Crystal structure of the N-terminal domain of Bqt4 in S.pombe | | Descriptor: | Bouquet formation protein 4 | | Authors: | Hu, C, Chen, Y. | | Deposit date: | 2017-09-05 | | Release date: | 2018-09-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | The Inner Nuclear Membrane Protein Bqt4 in Fission Yeast Contains a DNA-Binding Domain Essential for Telomere Association with the Nuclear Envelope.
Structure, 27, 2019
|
|
7TR6
 
 | | Cascade complex from type I-A CRISPR-Cas system | | Descriptor: | Cas11a, Cas5a, Cas7a, ... | | Authors: | Hu, C, Ni, D, Nam, K.H, Majumdar, S, McLean, J, Stahlberg, H, Terns, M, Ke, A. | | Deposit date: | 2022-01-28 | | Release date: | 2022-08-10 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Allosteric control of type I-A CRISPR-Cas3 complexes and establishment as effective nucleic acid detection and human genome editing tools.
Mol.Cell, 82, 2022
|
|
7TR8
 
 | | Cascade complex from type I-A CRISPR-Cas system | | Descriptor: | CRISPR-associated endonuclease Cas3-HD, CRISPR-associated helicase Cas3, Cas11a, ... | | Authors: | Hu, C, Ni, D, Nam, K.H, Majumdar, S, McLean, J, Stahlberg, H, Terns, M, Ke, A. | | Deposit date: | 2022-01-28 | | Release date: | 2022-08-10 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Allosteric control of type I-A CRISPR-Cas3 complexes and establishment as effective nucleic acid detection and human genome editing tools.
Mol.Cell, 82, 2022
|
|
7TRA
 
 | | Cascade complex from type I-A CRISPR-Cas system | | Descriptor: | CRISPR-associated endonuclease Cas3-HD, CRISPR-associated helicase Cas3, Cas11a, ... | | Authors: | Hu, C, Ni, D, Nam, K.H, Majumdar, S, McLean, J, Stahlberg, H, Terns, M, Ke, A. | | Deposit date: | 2022-01-28 | | Release date: | 2022-08-10 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Allosteric control of type I-A CRISPR-Cas3 complexes and establishment as effective nucleic acid detection and human genome editing tools.
Mol.Cell, 82, 2022
|
|
7TR9
 
 | | Cascade complex from type I-A CRISPR-Cas system | | Descriptor: | CRISPR-associated endonuclease Cas3-HD, CRISPR-associated helicase Cas3, Cas11a, ... | | Authors: | Hu, C, Ni, D, Nam, K.H, Majumdar, S, McLean, J, Stahlberg, H, Terns, M, Ke, A. | | Deposit date: | 2022-01-28 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Allosteric control of type I-A CRISPR-Cas3 complexes and establishment as effective nucleic acid detection and human genome editing tools.
Mol.Cell, 82, 2022
|
|
2MYP
 
 | |
2MYT
 
 | | An arsenate reductase in the intermediate state | | Descriptor: | Glutaredoxin arsenate reductase | | Authors: | Jin, C, Yu, C, Hu, C, Hu, Y. | | Deposit date: | 2015-01-30 | | Release date: | 2015-08-05 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | A Hybrid Mechanism for the Synechocystis Arsenate Reductase Revealed by Structural Snapshots during Arsenate Reduction.
J.Biol.Chem., 290, 2015
|
|
2MYN
 
 | | An arsenate reductase in reduced state | | Descriptor: | Glutaredoxin arsenate reductase | | Authors: | Jin, C, Yu, C, Hu, C, Hu, Y. | | Deposit date: | 2015-01-30 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A Hybrid Mechanism for the Synechocystis Arsenate Reductase Revealed by Structural Snapshots during Arsenate Reduction.
J.Biol.Chem., 290, 2015
|
|
