5F4L
 
 | | HIV-1 gp120 complex with JP-III-048 | | Descriptor: | ENVELOPE GLYCOPROTEIN GP120 of HIV-1 clade C, ~{N}'-[(1~{R},2~{R})-2-(carbamimidamidomethyl)-6-(methylaminomethyl)-2,3-dihydro-1~{H}-inden-1-yl]-~{N}-(4-chloranyl-3-fluoranyl-phenyl)ethanediamide | | Authors: | Liang, S, Hendrickson, W.A. | | Deposit date: | 2015-12-03 | | Release date: | 2016-03-30 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Small-Molecule CD4-Mimics: Structure-Based Optimization of HIV-1 Entry Inhibition.
Acs Med.Chem.Lett., 7, 2016
|
|
2FIT
 
 | | FHIT (FRAGILE HISTIDINE TRIAD PROTEIN) | | Descriptor: | FRAGILE HISTIDINE PROTEIN, SULFATE ION, beta-D-fructofuranose | | Authors: | Lima, C.D, D'Amico, K.L, Naday, I, Rosenbaum, G, Westbrook, E.M, Hendrickson, W.A. | | Deposit date: | 1997-05-17 | | Release date: | 1997-11-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | MAD analysis of FHIT, a putative human tumor suppressor from the HIT protein family.
Structure, 5, 1997
|
|
2HB5
 
 | | Crystal Structure of the Moloney Murine Leukemia Virus RNase H Domain | | Descriptor: | MAGNESIUM ION, Reverse transcriptase/ribonuclease H, SULFATE ION | | Authors: | Lim, D, Gregorio, G.G, Bingman, C.A, Martinez-Hackert, E, Hendrickson, W.A, Goff, S.P. | | Deposit date: | 2006-06-13 | | Release date: | 2006-08-29 | | Last modified: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Crystal Structure of the Moloney Murine Leukemia Virus RNase H Domain.
J.Virol., 80, 2006
|
|
6VX7
 
 | | bestrophin-2 Ca2+-bound state (5 mM Ca2+) | | Descriptor: | Bestrophin, CALCIUM ION, CHLORIDE ION | | Authors: | Owji, A.P, Zhao, Q, Ji, C, Kittredge, A, Hopiavuori, A, Fu, Z, Ward, N, Clarke, O, Shen, Y, Zhang, Y, Hendrickson, W.A, Yang, T. | | Deposit date: | 2020-02-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.36 Å) | | Cite: | Structural and functional characterization of the bestrophin-2 anion channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VX8
 
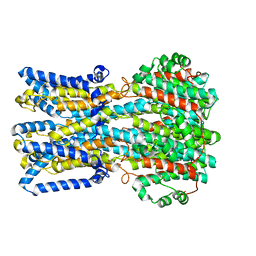 | | bestrophin-2 Ca2+- unbound state 2 (EGTA only) | | Descriptor: | Bestrophin, CHLORIDE ION | | Authors: | Owji, A.P, Zhao, Q, Ji, C, Kittredge, A, Hopiavuori, A, Fu, Z, Ward, N, Clarke, O, Shen, Y, Zhang, Y, Hendrickson, W.A, Yang, T. | | Deposit date: | 2020-02-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Structural and functional characterization of the bestrophin-2 anion channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VX6
 
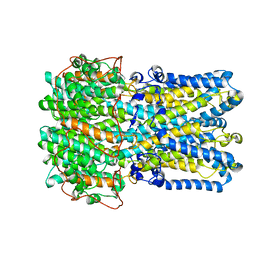 | | bestrophin-2 Ca2+-bound state (250 nM Ca2+) | | Descriptor: | Bestrophin, CALCIUM ION, CHLORIDE ION | | Authors: | Owji, A.P, Zhao, Q, Ji, C, Kittredge, A, Hopiavuori, A, Fu, Z, Ward, N, Clarke, O, Shen, Y, Zhang, Y, Hendrickson, W.A, Yang, T. | | Deposit date: | 2020-02-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and functional characterization of the bestrophin-2 anion channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VX9
 
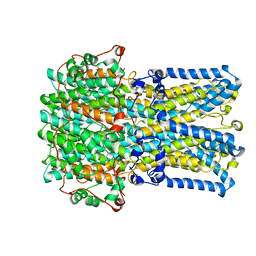 | | bestrophin-2 Ca2+- unbound state 1 (EGTA only) | | Descriptor: | Bestrophin, CHLORIDE ION | | Authors: | Owji, A.P, Zhao, Q, Ji, C, Kittredge, A, Hopiavuori, A, Fu, Z, Ward, N, Clarke, O, Shen, Y, Zhang, Y, Hendrickson, W.A, Yang, T. | | Deposit date: | 2020-02-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.17 Å) | | Cite: | Structural and functional characterization of the bestrophin-2 anion channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VX5
 
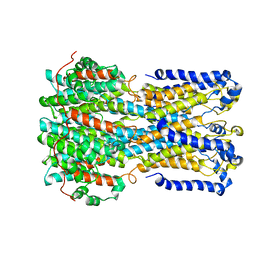 | | bestrophin-2 Ca2+- unbound state (250 nM Ca2+) | | Descriptor: | Bestrophin, CHLORIDE ION | | Authors: | Owji, A.P, Zhao, Q, Ji, C, Kittredge, A, Hopiavuori, A, Fu, Z, Ward, N, Clarke, O, Shen, Y, Zhang, Y, Hendrickson, W.A, Yang, T. | | Deposit date: | 2020-02-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structural and functional characterization of the bestrophin-2 anion channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6WIV
 
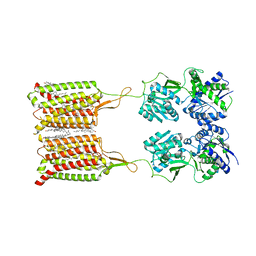 | | Structure of human GABA(B) receptor in an inactive state | | Descriptor: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-{[(9Z)-octadec-9-enoyl]oxy}propyl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Park, J, Fu, Z, Frangaj, A, Liu, J, Mosyak, L, Shen, T, Slavkovich, V.N, Ray, K.M, Taura, J, Cao, B, Geng, Y, Zuo, H, Kou, Y, Grassucci, R, Chen, S, Liu, Z, Lin, X, Williams, J.P, Rice, W.J, Eng, E.T, Huang, R.K, Soni, R.K, Kloss, B, Yu, Z, Javitch, J.A, Hendrickson, W.A, Slesinger, P.A, Quick, M, Graziano, J, Yu, H, Fiehn, O, Clarke, O.B, Frank, J, Fan, Q.R. | | Deposit date: | 2020-04-10 | | Release date: | 2020-07-01 | | Last modified: | 2020-08-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of human GABABreceptor in an inactive state.
Nature, 584, 2020
|
|
6IZ5
 
 | |
6IZ6
 
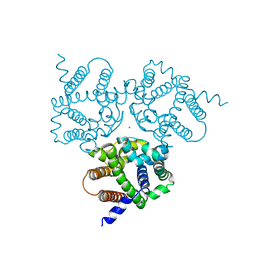 | | Crystal Structure Analysis of TRIC counter-ion channels in calcium release | | Descriptor: | CALCIUM ION, Trimeric intracellular cation channel type B-B | | Authors: | Liu, X.L, Wang, X.H, Su, M, Hendrickson, W.A, Chen, Y.H. | | Deposit date: | 2018-12-18 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.293 Å) | | Cite: | Structural basis for activity of TRIC counter-ion channels in calcium release.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6IZF
 
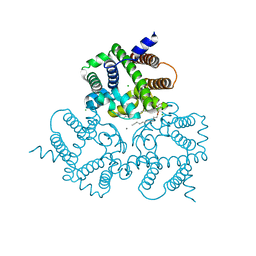 | | Structural basis for activity of TRIC counter-ion channels in calcium release | | Descriptor: | (2R)-1-(dodecanoyloxy)-3-hydroxypropan-2-yl (5E,8E,11E)-tetradeca-5,8,11-trienoate, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wang, X.H, Zeng, Y, Su, M, Hendrickson, W.A, Chen, Y.H. | | Deposit date: | 2018-12-19 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for activity of TRIC counter-ion channels in calcium release.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6IZ4
 
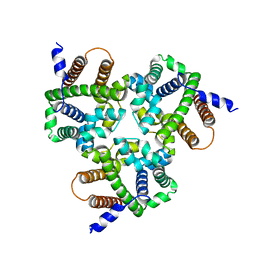 | | Crystal Structure Analysis of TRIC counter-ion channels in calcium release | | Descriptor: | Trimeric intracellular cation channel type B-B | | Authors: | Wang, X.H, Zeng, Y, Gao, F, Su, M, Hendrickson, W.A, Chen, Y.H. | | Deposit date: | 2018-12-18 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.098 Å) | | Cite: | Structural basis for activity of TRIC counter-ion channels in calcium release.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
1BUN
 
 | | STRUCTURE OF BETA2-BUNGAROTOXIN: POTASSIUM CHANNEL BINDING BY KUNITZ MODULES AND TARGETED PHOSPHOLIPASE ACTION | | Descriptor: | BETA2-BUNGAROTOXIN, SODIUM ION | | Authors: | Kwong, P.D, Mcdonald, N.Q, Sigler, P.B, Hendrickson, W.A. | | Deposit date: | 1995-10-15 | | Release date: | 1996-04-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of beta 2-bungarotoxin: potassium channel binding by Kunitz modules and targeted phospholipase action.
Structure, 3, 1995
|
|
1CDJ
 
 | | STRUCTURE OF T-CELL SURFACE GLYCOPROTEIN CD4 | | Descriptor: | T-CELL SURFACE GLYCOPROTEIN CD4 | | Authors: | Wu, H, Myszka, D, Tendian, S.W, Brouillette, C.G, Sweet, R.W, Chaiken, I.M, Hendrickson, W.A. | | Deposit date: | 1996-11-11 | | Release date: | 1997-04-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Kinetic and structural analysis of mutant CD4 receptors that are defective in HIV gp120 binding.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1CDH
 
 | |
4I54
 
 | |
4I53
 
 | |
4JNE
 
 | | Allosteric opening of the polypeptide-binding site when an Hsp70 binds ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, Hsp70 CHAPERONE DnaK, ... | | Authors: | Qi, R, Sarbeng, E.B, Liu, Q, Le, K.Q, Xu, X, Xu, H, Yang, J, Wong, J.L, Vorvis, C, Hendrickson, W.A, Zhou, L, Liu, Q. | | Deposit date: | 2013-03-15 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Allosteric opening of the polypeptide-binding site when an Hsp70 binds ATP.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4JNF
 
 | | Allosteric opening of the polypeptide-binding site when an Hsp70 binds ATP | | Descriptor: | Hsp70 CHAPERONE DnaK | | Authors: | Qi, R, Sarbeng, E.B, Liu, Q, Le, K.Q, Xu, X, Xu, H, Yang, J, Wong, J.L, Vorvis, C, Hendrickson, W.A, Zhou, L, Liu, Q. | | Deposit date: | 2013-03-15 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.621 Å) | | Cite: | Allosteric opening of the polypeptide-binding site when an Hsp70 binds ATP.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4JN4
 
 | | Allosteric opening of the polypeptide-binding site when an Hsp70 binds ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chaperone protein DnaK, GLYCEROL, ... | | Authors: | Qi, R, Sarbeng, E.B, Liu, Q, Le, K.Q, Xu, X, Xu, H, Yang, J, Wong, J.L, Vorvis, C, Hendrickson, W.A, Zhou, L, Liu, Q. | | Deposit date: | 2013-03-14 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Allosteric opening of the polypeptide-binding site when an Hsp70 binds ATP.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4RYQ
 
 | | Crystal structure of BcTSPO, type 2 at 1.7 Angstrom | | Descriptor: | Integral membrane protein, [(Z)-octadec-9-enyl] (2R)-2,3-bis(oxidanyl)propanoate | | Authors: | Guo, Y, Liu, Q, Hendrickson, W.A, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2014-12-16 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Protein structure. Structure and activity of tryptophan-rich TSPO proteins.
Science, 347, 2015
|
|
4RYM
 
 | |
4TKR
 
 | | Native-SAD phasing for ThiT from Listeria monocytogenes serovar. | | Descriptor: | 2-O-octyl-beta-D-glucopyranose, THIAMINE DIPHOSPHATE, Thiamine transporter thia | | Authors: | Guo, Y, Liu, Q, Hendrickson, W.A, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2014-05-27 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.0023 Å) | | Cite: | Multi-crystal native SAD analysis at 6 keV.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4RYJ
 
 | |
