3PH0
 
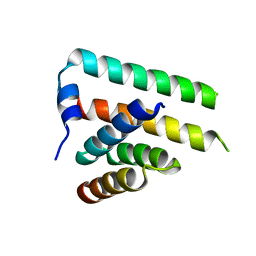 | | Crystal structure of the heteromolecular chaperone, AscE-AscG, from the type III secretion system in Aeromonas hydrophila | | Descriptor: | AscE, AscG | | Authors: | Chatterjee, C, Kumar, S, Chakraborty, S, Tan, Y.W, Leung, K.Y, Sivaraman, J, Mok, Y.K. | | Deposit date: | 2010-11-03 | | Release date: | 2011-07-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the heteromolecular chaperone, AscE-AscG, from the type III secretion system in Aeromonas hydrophila
Plos One, 6, 2011
|
|
1B8T
 
 | | SOLUTION STRUCTURE OF THE CHICKEN CRP1 | | Descriptor: | PROTEIN (CRP1), ZINC ION | | Authors: | Yao, X, Perez-Alvarado, G.C, Louis, H.A, Pomies, P, Hatt, C, Summers, M.F, Beckerle, M.C. | | Deposit date: | 1999-02-02 | | Release date: | 1999-05-06 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the chicken cysteine-rich protein, CRP1, a double-LIM protein implicated in muscle differentiation.
Biochemistry, 38, 1999
|
|
3J2U
 
 | | Kinesin-13 KLP10A HD in complex with CS-tubulin and a microtubule | | Descriptor: | Kinesin-like protein Klp10A, Tubulin alpha-1A chain, Tubulin beta-2B chain | | Authors: | Asenjo, A.B, Chatterjee, C, Tan, D, DePaoli, V, Rice, W.J, Diaz-Avalos, R, Silvestry, M, Sosa, H. | | Deposit date: | 2013-01-10 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (10.8 Å) | | Cite: | Structural model for tubulin recognition and deformation by Kinesin-13 microtubule depolymerases.
Cell Rep, 3, 2013
|
|
6UGM
 
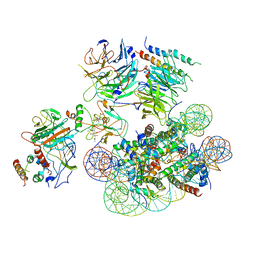 | | Structural basis of COMPASS eCM recognition of an unmodified nucleosome | | Descriptor: | Bre2, DNA (146-MER), H3 N-terminus, ... | | Authors: | Hsu, P.L, Shi, H, Zheng, N. | | Deposit date: | 2019-09-26 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural Basis of H2B Ubiquitination-Dependent H3K4 Methylation by COMPASS.
Mol.Cell, 76, 2019
|
|
4YK0
 
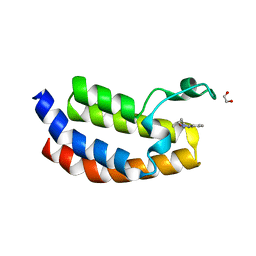 | | Crystal structure of the CBP bromodomain in complex with CPI098 | | Descriptor: | (4R)-4-methyl-1,3,4,5-tetrahydro-2H-1,5-benzodiazepin-2-one, 1,2-ETHANEDIOL, CREB-binding protein | | Authors: | Bellon, S.F, Jayaram, H. | | Deposit date: | 2015-03-03 | | Release date: | 2016-04-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Regulatory T Cell Modulation by CBP/EP300 Bromodomain Inhibition.
J.Biol.Chem., 291, 2016
|
|
6GKI
 
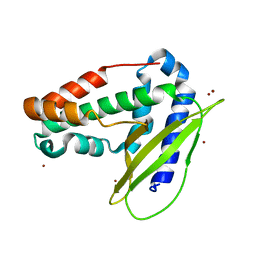 | | Structure of E coli MlaC in Variously Loaded States | | Descriptor: | BROMIDE ION, GLYCEROL, Probable phospholipid-binding protein MlaC | | Authors: | Knowles, T.J, Lovering, A.L. | | Deposit date: | 2018-05-21 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Evidence for phospholipid export from the bacterial inner membrane by the Mla ABC transport system.
Nat Microbiol, 4, 2019
|
|
9F1Z
 
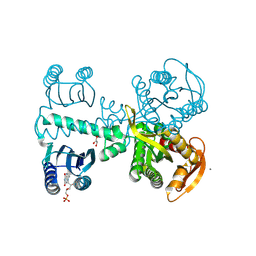 | | Crystal structure of adenylyate cyclase variant Y6W | | Descriptor: | CALCIUM ION, FLAVIN MONONUCLEOTIDE, Family 3 adenylate cyclase, ... | | Authors: | Kapetanaki, S, Coquelle, N, Weik, M. | | Deposit date: | 2024-04-20 | | Release date: | 2025-02-26 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of a bacterial photoactivated adenylate cyclase determined by serial femtosecond and serial synchrotron crystallography.
Iucrj, 11, 2024
|
|
8E90
 
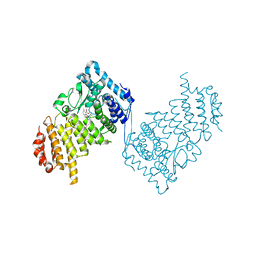 | | Inhibition of Human Menin by SNDX-5613 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-({4-[7-({(1r,4r)-4-[(ethanesulfonyl)amino]cyclohexyl}methyl)-2,7-diazaspiro[3.5]nonan-2-yl]pyrimidin-5-yl}oxy)-N-ethyl-5-fluoro-N-(propan-2-yl)benzamide, ... | | Authors: | McKeever, B.M, KULKARNI, S, McGeehan, G.M. | | Deposit date: | 2022-08-26 | | Release date: | 2022-12-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | MEN1 mutations mediate clinical resistance to menin inhibition.
Nature, 615, 2023
|
|
8P82
 
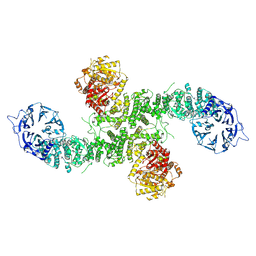 | | Cryo-EM structure of dimeric UBR5 | | Descriptor: | E3 ubiquitin-protein ligase UBR5, ZINC ION | | Authors: | Aguirre, J.D, Kater, L, Kempf, G, Cavadini, S, Thoma, N.H. | | Deposit date: | 2023-05-31 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | UBR5 forms ligand-dependent complexes on chromatin to regulate nuclear hormone receptor stability.
Mol.Cell, 83, 2023
|
|
8P83
 
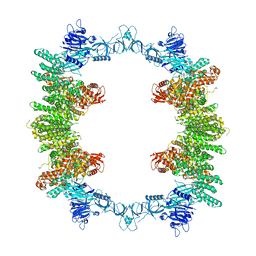 | | Cryo-EM structure of full-length human UBR5 (homotetramer) | | Descriptor: | E3 ubiquitin-protein ligase UBR5 | | Authors: | Aguirre, J.D, Kater, L, Kempf, G, Cavadini, S, Thoma, N.H. | | Deposit date: | 2023-05-31 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | UBR5 forms ligand-dependent complexes on chromatin to regulate nuclear hormone receptor stability.
Mol.Cell, 83, 2023
|
|
7UJ4
 
 | | Inhibition of Human Menin by SNDX-5613 | | Descriptor: | 2-({4-[7-({(1r,4r)-4-[(ethanesulfonyl)amino]cyclohexyl}methyl)-2,7-diazaspiro[3.5]nonan-2-yl]pyrimidin-5-yl}oxy)-N-ethyl-5-fluoro-N-(propan-2-yl)benzamide, Isoform 2 of Menin, MAGNESIUM ION | | Authors: | McKeever, B.M, Kulkarni, S, McGeehan, G.M. | | Deposit date: | 2022-03-30 | | Release date: | 2022-12-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | MEN1 mutations mediate clinical resistance to menin inhibition.
Nature, 615, 2023
|
|
6UH5
 
 | | Structural basis of COMPASS eCM recognition of the H2Bub nucleosome | | Descriptor: | Bre2, DNA (146-MER), H3 N-terminus, ... | | Authors: | Hsu, P.L, Shi, H, Zheng, N. | | Deposit date: | 2019-09-26 | | Release date: | 2019-11-20 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural Basis of H2B Ubiquitination-Dependent H3K4 Methylation by COMPASS.
Mol.Cell, 76, 2019
|
|
6CHG
 
 | | Crystal structure of the yeast COMPASS catalytic module | | Descriptor: | H3, Histone-lysine N-methyltransferase, H3 lysine-4 specific, ... | | Authors: | Hsu, P.L, Li, H, Zheng, N. | | Deposit date: | 2018-02-22 | | Release date: | 2018-08-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.985 Å) | | Cite: | Crystal Structure of the COMPASS H3K4 Methyltransferase Catalytic Module.
Cell, 174, 2018
|
|
5K1A
 
 | |
5K1C
 
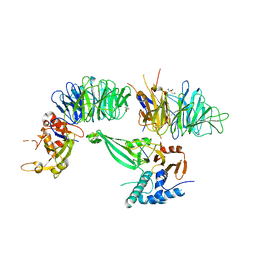 | | Crystal structure of the UAF1/WDR20/USP12 complex | | Descriptor: | PHOSPHATE ION, TRIS(HYDROXYETHYL)AMINOMETHANE, Ubiquitin carboxyl-terminal hydrolase 12, ... | | Authors: | Li, H, D'Andrea, A.D, Zheng, N. | | Deposit date: | 2016-05-18 | | Release date: | 2016-07-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Allosteric Activation of Ubiquitin-Specific Proteases by beta-Propeller Proteins UAF1 and WDR20.
Mol.Cell, 63, 2016
|
|
5K16
 
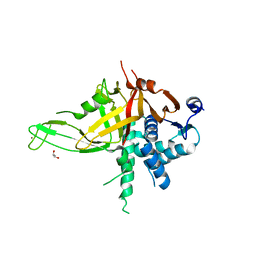 | |
5K1B
 
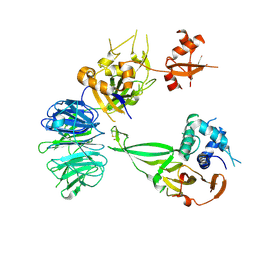 | |
5DBM
 
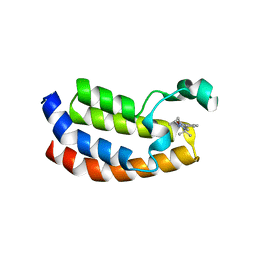 | | Crystal structure of the CBP bromodomain in complex with CPI703 | | Descriptor: | (4R)-6-(1-tert-butyl-1H-pyrazol-4-yl)-4-methyl-1,3,4,5-tetrahydro-2H-1,5-benzodiazepin-2-one, CREB-binding protein | | Authors: | Setser, J.W, Poy, F, Bellon, S.F. | | Deposit date: | 2015-08-21 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Regulatory T Cell Modulation by CBP/EP300 Bromodomain Inhibition.
J.Biol.Chem., 291, 2016
|
|
6PKC
 
 | | Inhibition of Human Menin by VTP-50469 | | Descriptor: | 5-fluoro-2-({4-[7-({trans-4-[(methylsulfonyl)amino]cyclohexyl}methyl)-2,7-diazaspiro[3.5]nonan-2-yl]pyrimidin-5-yl}oxy)-N,N-di(propan-2-yl)benzamide, Menin,Menin | | Authors: | McKeever, B.M, Chen, G, Van Orton, R. | | Deposit date: | 2019-06-29 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Menin-MLL Inhibitor Induces Specific Chromatin Changes and Eradicates Disease in Models of MLL-Rearranged Leukemia.
Cancer Cell, 36, 2019
|
|
5K19
 
 | |
2MD3
 
 | |
2MD4
 
 | |
2MD2
 
 | |
2MD1
 
 | |
7JZV
 
 | | Cryo-EM structure of the BRCA1-UbcH5c/BARD1 E3-E2 module bound to a nucleosome | | Descriptor: | BRCA1,Ubiquitin-conjugating enzyme E2 D3, BRCA1-associated RING domain protein 1, Histone H2A type 2-A, ... | | Authors: | Witus, S.R, Burrell, A.L, Hansen, J.M, Farrell, D.P, Dimaio, F, Kollman, J.M, Klevit, R.E. | | Deposit date: | 2020-09-02 | | Release date: | 2021-02-17 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | BRCA1/BARD1 site-specific ubiquitylation of nucleosomal H2A is directed by BARD1.
Nat.Struct.Mol.Biol., 28, 2021
|
|
