2ZO2
 
 | | Mouse NP95 SRA domain non-specific DNA complex | | Descriptor: | DNA (5'-D(*DAP*DAP*DCP*DTP*DGP*DCP*DGP*DCP*DAP*DGP*DTP*DT)-3'), E3 ubiquitin-protein ligase UHRF1, PHOSPHATE ION | | Authors: | Hashimoto, H, Horton, J.R, Cheng, X. | | Deposit date: | 2008-05-05 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix
Nature, 455, 2008
|
|
2ZO1
 
 | | Mouse NP95 SRA domain DNA specific complex 2 | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*DGP*DTP*DCP*DAP*DGP*(5CM)P*DGP*DCP*DAP*DAP*DTP*DGP*DG)-3'), DNA (5'-D(*DTP*DCP*DCP*DAP*DTP*DGP*DCP*DGP*DCP*DTP*DGP*DAP*DC)-3'), ... | | Authors: | Hashimoto, H, Horton, J.R, Cheng, X. | | Deposit date: | 2008-05-05 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix
Nature, 455, 2008
|
|
2ZO0
 
 | | mouse NP95 SRA domain DNA specific complex 1 | | Descriptor: | DNA (5'-D(*DGP*DTP*DCP*DAP*DGP*(5CM)P*DGP*DCP*DAP*DAP*DTP*DGP*DG)-3'), DNA (5'-D(*DTP*DCP*DCP*DAP*DTP*DGP*DCP*DGP*DCP*DTP*DGP*DAP*DC)-3'), E3 ubiquitin-protein ligase UHRF1 | | Authors: | Hashimoto, H, Horton, J.R, Cheng, X. | | Deposit date: | 2008-05-05 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix
Nature, 455, 2008
|
|
3ALR
 
 | | Crystal structure of Nanos | | Descriptor: | Nanos protein, ZINC ION | | Authors: | Hashimoto, H, Hara, K, Hishiki, A, Kawaguchi, S, Shichijo, N, Nakamura, K, Unzai, S, Tamaru, Y, Shimizu, T, Sato, M. | | Deposit date: | 2010-08-06 | | Release date: | 2011-02-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of zinc-finger domain of Nanos and its functional implications
Embo Rep., 11, 2010
|
|
3AYU
 
 | | Crystal structure of MMP-2 active site mutant in complex with APP-drived decapeptide inhibitor | | Descriptor: | 72 kDa type IV collagenase, Amyloid beta A4 protein, CALCIUM ION, ... | | Authors: | Hashimoto, H, Takeuchi, T, Komatsu, K, Miyazaki, K, Sato, M, Higashi, S. | | Deposit date: | 2011-05-17 | | Release date: | 2011-08-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for matrix metalloproteinase-2 (MMP-2)-selective inhibitory action of {beta}-amyloid precursor protein-derived inhibitor
J.Biol.Chem., 2011
|
|
5YD8
 
 | |
3W08
 
 | | Crystal structure of aldoxime dehydratase | | Descriptor: | Aldoxime dehydratase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hashimoto, H, Nomura, J, Hashimoto, Y, Oinuma, K.I, Wada, K, Hishiki, A, Hara, K, Kobayashi, M. | | Deposit date: | 2012-10-24 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of aldoxime dehydratase and its catalytic mechanism involved in carbon-nitrogen triple-bond synthesis.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3VKX
 
 | | Structure of PCNA | | Descriptor: | 3,5,3'TRIIODOTHYRONINE, CHLORIDE ION, Proliferating cell nuclear antigen, ... | | Authors: | Hashimoto, H, Hishiki, A, Shimizu, T, Sato, M, Punchihewa, C, Connelly, M, Actis, M, Waddell, B, Pagala, V, Fujii, N. | | Deposit date: | 2011-11-26 | | Release date: | 2012-03-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of small molecule proliferating cell nuclear antigen (PCNA) inhibitor that disrupts interactions with PIP-box proteins and inhibits DNA replication
J.Biol.Chem., 287, 2012
|
|
3WGW
 
 | | Structure of PCNA bound to a small molecule inhibitor | | Descriptor: | 4-{4-[(2S)-2-amino-3-hydroxypropyl]-2,6-diiodophenoxy}phenol, Proliferating cell nuclear antigen, SULFATE ION | | Authors: | Hashimoto, H. | | Deposit date: | 2013-08-12 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A small molecule inhibitor of monoubiquitinated Proliferating Cell Nuclear Antigen (PCNA) inhibits repair of interstrand DNA cross-link, enhances DNA double strand break, and sensitizes cancer cells to cisplatin.
J.Biol.Chem., 289, 2014
|
|
3A57
 
 | | Crystal structure of Thermostable Direct Hemolysin | | Descriptor: | Thermostable direct hemolysin 2 | | Authors: | Hashimoto, H, Yanagihara, I, Nakahira, K, Hamada, D, Ikegami, T, Mayanagi, K, Kaieda, S, Fukui, T, Ohnishi, K, Kajiyama, S, Yamane, T, Ikeguchi, M, Honda, T, Shimizu, T, Sato, M. | | Deposit date: | 2009-08-03 | | Release date: | 2010-03-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and functional characterization of Vibrio parahaemolyticus thermostable direct hemolysin
J.Biol.Chem., 285, 2010
|
|
4XZG
 
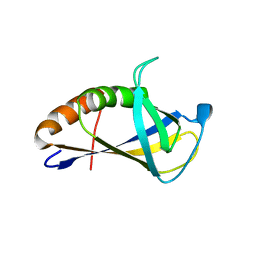 | | Crystal structure of HIRAN domain of human HLTF | | Descriptor: | Helicase-like transcription factor | | Authors: | Ikegaya, Y, Hara, K, Hashimoto, H. | | Deposit date: | 2015-02-04 | | Release date: | 2015-04-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a Novel DNA-binding Domain of Helicase-like Transcription Factor (HLTF) and Its Functional Implication in DNA Damage Tolerance
J.Biol.Chem., 290, 2015
|
|
8W7D
 
 | | Crystal structure of EcPPAT-FR901483 complex | | Descriptor: | Amidophosphoribosyltransferase, [(1S,3S,6S,7S,8R,9S)-6-[(4-methoxyphenyl)methyl]-3-(methylamino)-7-oxidanyl-5-azatricyclo[6.3.1.0^1,5]dodecan-9-yl] dihydrogen phosphate | | Authors: | Hara, K, Hashimoto, H, Nakahara, M, Sato, M, Watanabe, K. | | Deposit date: | 2023-08-30 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Uncommon Arrangement of Self-resistance Allows Biosynthesis of de novo Purine Biosynthesis Inhibitor that Acts as an Immunosuppressor.
J.Am.Chem.Soc., 145, 2023
|
|
8WU8
 
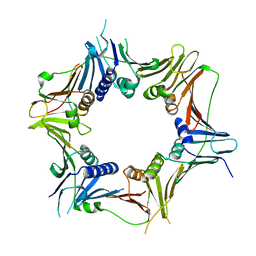 | | Crystal structure of the human RAD9-RAD1(F64A/M256A/F266A)-HUS1-RHINO(88-99) complex | | Descriptor: | Cell cycle checkpoint control protein RAD9A, Cell cycle checkpoint protein RAD1, Checkpoint protein HUS1, ... | | Authors: | Hara, K, Nagata, K, Iida, N, Hashimoto, H. | | Deposit date: | 2023-10-20 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structural basis for intra- and intermolecular interactions on RAD9 subunit of 9-1-1 checkpoint clamp implies functional 9-1-1 regulation by RHINO.
J.Biol.Chem., 300, 2024
|
|
8Y2S
 
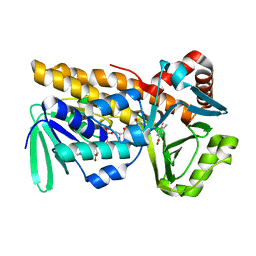 | | P-hydroxybenzoate hydroxylase complexed with 4-hydroxy-3-methylbenzoic acid | | Descriptor: | 3-methyl-4-oxidanyl-benzoic acid, 4-hydroxybenzoate 3-monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Hara, K, Hashimoto, H, Matsushita, T, Kishimoto, S, Watanabe, K. | | Deposit date: | 2024-01-27 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional Enhancement of Flavin-Containing Monooxygenase through Machine Learning Methodology
Acs Catalysis, 14, 2024
|
|
7FI3
 
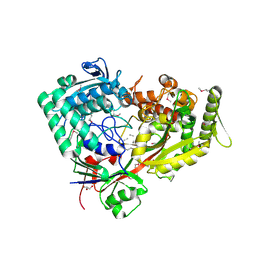 | | Archaeal oligopeptide permease A (OppA) from Thermococcus kodakaraensis in complex with an endogenous pentapeptide | | Descriptor: | ABC-type dipeptide/oligopeptide transport system, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | Authors: | Yokoyama, H, Kamei, N, Konishi, K, Hara, K, Hashimoto, H. | | Deposit date: | 2021-07-30 | | Release date: | 2022-04-13 | | Last modified: | 2022-06-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for peptide recognition by archaeal oligopeptide permease A.
Proteins, 90, 2022
|
|
8FJN
 
 | |
8FJM
 
 | |
8GNN
 
 | | Crystal structure of the human RAD9-RAD1-HUS1-RAD17 complex | | Descriptor: | Cell cycle checkpoint control protein RAD9A, Cell cycle checkpoint protein RAD1, Cell cycle checkpoint protein RAD17, ... | | Authors: | Hara, K, Nagata, K, Iida, N, Hashimoto, H. | | Deposit date: | 2022-08-24 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.119 Å) | | Cite: | The 9-1-1 DNA clamp subunit RAD1 forms specific interactions with clamp loader RAD17, revealing functional implications for binding-protein RHINO.
J.Biol.Chem., 299, 2023
|
|
5H6K
 
 | | DNA targeting ADP-ribosyltransferase Pierisin-1 | | Descriptor: | 1,2-ETHANEDIOL, Pierisin-1 | | Authors: | Oda, T, Hirabayashi, H, Shikauchi, G, Takamura, R, Hiraga, K, Minami, H, Hashimoto, H, Yamamoto, M, Wakabayashi, K, Sugimura, T, Shimizu, T, Sato, M. | | Deposit date: | 2016-11-14 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of autoinhibition and activation of the DNA-targeting ADP-ribosyltransferase pierisin-1
J. Biol. Chem., 292, 2017
|
|
5H6N
 
 | | DNA targeting ADP-ribosyltransferase Pierisin-1, autoinhibitory form | | Descriptor: | Pierisin-1 | | Authors: | Oda, T, Hirabayashi, H, Shikauchi, G, Takamura, R, Hiraga, K, Minami, H, Hashimoto, H, Yamamoto, M, Wakabayashi, K, Sugimura, T, Shimizu, T, Sato, M. | | Deposit date: | 2016-11-14 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of autoinhibition and activation of the DNA-targeting ADP-ribosyltransferase pierisin-1
J. Biol. Chem., 292, 2017
|
|
5FHI
 
 | | Crystallographic structure of PsoE without Co | | Descriptor: | GLUTATHIONE, Glutathione S-transferase, putative | | Authors: | Hara, K, Hashimoto, H, Yamamoto, T, Tsunematsu, Y, Watanabe, K. | | Deposit date: | 2015-12-22 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Oxidative trans to cis Isomerization of Olefins in Polyketide Biosynthesis.
Angew. Chem. Int. Ed. Engl., 55, 2016
|
|
5F8B
 
 | | Crystallographic Structure of PsoE with Co | | Descriptor: | (5~{S},8~{S},9~{R})-2-[(~{E})-hex-1-enyl]-8-methoxy-3-methyl-9-oxidanyl-8-(phenylcarbonyl)-1-oxa-7-azaspiro[4.4]non-2-ene-4,6-dione, COBALT (II) ION, GLUTATHIONE, ... | | Authors: | Hara, K, Hashimoto, H, Yamamoto, T, Tsunematsu, Y, Watanabe, K. | | Deposit date: | 2015-12-09 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Oxidative trans to cis Isomerization of Olefins in Polyketide Biosynthesis.
Angew. Chem. Int. Ed. Engl., 55, 2016
|
|
3I4D
 
 | | Photosynthetic reaction center from rhodobacter sphaeroides 2.4.1 | | Descriptor: | (2R,3S)-heptane-1,2,3-triol, 1,4-DIETHYLENE DIOXIDE, BACTERIOCHLOROPHYLL A, ... | | Authors: | Fujii, R, Adachi, S, Roszak, A.W, Gardiner, A.T, Cogdell, R.J, Isaacs, N.W, Koshihara, S, Hashimoto, H. | | Deposit date: | 2009-07-01 | | Release date: | 2010-12-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure of the carotenoid bound to the reaction centre from Rhodobacter sphaeroides 2.4.1 revealed by time-resolved X-ray crystallography
To be Published
|
|
3PUS
 
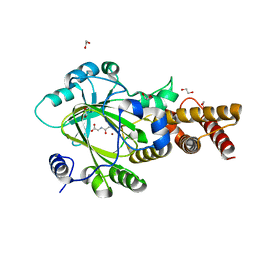 | | PHF2 Jumonji-NOG-Ni(II) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, N-OXALYLGLYCINE, ... | | Authors: | Horton, J.R, Upadhyay, A.K, Hashimoto, H, Zhang, X, Cheng, X. | | Deposit date: | 2010-12-06 | | Release date: | 2011-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural basis for human PHF2 Jumonji domain interaction with metal ions.
J.Mol.Biol., 406, 2011
|
|
3PU8
 
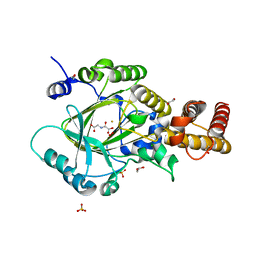 | | PHF2 Jumonji-NOG-Fe(II) complex | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FE (III) ION, ... | | Authors: | Horton, J.R, Upadhyay, A.K, Hashimoto, H, Zhang, X, Cheng, X. | | Deposit date: | 2010-12-03 | | Release date: | 2011-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.943 Å) | | Cite: | Structural basis for human PHF2 Jumonji domain interaction with metal ions.
J.Mol.Biol., 406, 2011
|
|
