1NUT
 
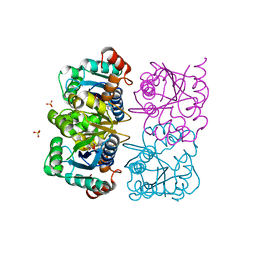 | | CRYSTAL STRUCTURE OF HUMAN CYTOSOLIC NMN/NaMN ADENYLYLTRANSFERASE COMPLEXED WITH ATP ANALOG | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, FKSG76, SULFATE ION | | Authors: | Zhang, X, Kurnasov, O.V, Karthikeyan, S, Grishin, N.V, Osterman, A.L, Zhang, H. | | Deposit date: | 2003-02-01 | | Release date: | 2003-06-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | STRUCTURAL CHARACTERIZATION OF A HUMAN CYTOSOLIC NMN/NaMN
ADENYLYLTRANSFERASE AND IMPLICATION IN HUMAN NAD BIOSYNTHESIS
J.Biol.Chem., 278, 2003
|
|
1NUQ
 
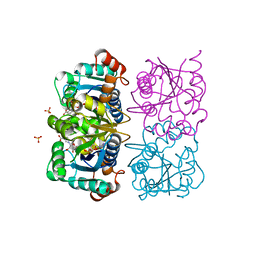 | | CRYSTAL STRUCTURE OF HUMAN CYTOSOLIC NMN/NaMN ADENYLYLTRANSFERASE COMPLEXED WITH NaAD | | Descriptor: | FKSG76, NICOTINIC ACID ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Zhang, X, Kurnasov, O.V, Karthikeyan, S, Grishin, N.V, Osterman, A.L, Zhang, H. | | Deposit date: | 2003-02-01 | | Release date: | 2003-06-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | STRUCTURAL CHARACTERIZATION OF A HUMAN CYTOSOLIC NMN/NaMN
ADENYLYLTRANSFERASE AND IMPLICATION IN HUMAN NAD BIOSYNTHESIS
J.Biol.Chem., 278, 2003
|
|
5TEC
 
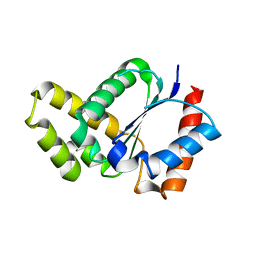 | | Crystal structure of the TIR domain from the Arabidopsis thaliana NLR protein SNC1 | | Descriptor: | Protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1 | | Authors: | Zhang, X, Bentham, A, Ve, T, Williams, S.J, Kobe, B. | | Deposit date: | 2016-09-20 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Multiple functional self-association interfaces in plant TIR domains.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1NUS
 
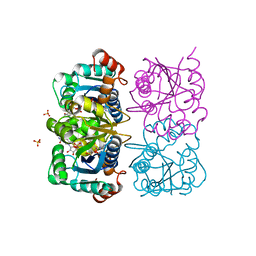 | | CRYSTAL STRUCTURE OF HUMAN CYTOSOLIC NMN/NaMN ADENYLYLTRANSFERASE COMPLEXED WITH ATP ANALOG AND NMN | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, FKSG76, ... | | Authors: | Zhang, X, Kurnasov, O.V, Karthikeyan, S, Grishin, N.V, Osterman, A.L, Zhang, H. | | Deposit date: | 2003-02-01 | | Release date: | 2003-06-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | STRUCTURAL CHARACTERIZATION OF A HUMAN CYTOSOLIC NMN/NaMN
ADENYLYLTRANSFERASE AND IMPLICATION IN HUMAN NAD BIOSYNTHESIS
J.Biol.Chem., 278, 2003
|
|
1NUU
 
 | | CRYSTAL STRUCTURE OF HUMAN CYTOSOLIC NMN/NaMN ADENYLYLTRANSFERASE COMPLEXED WITH NAD | | Descriptor: | FKSG76, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Zhang, X, Kurnasov, O.V, Karthikeyan, S, Grishin, N.V, Osterman, A.L, Zhang, H. | | Deposit date: | 2003-02-01 | | Release date: | 2003-06-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | STRUCTURAL CHARACTERIZATION OF A HUMAN CYTOSOLIC NMN/NaMN
ADENYLYLTRANSFERASE AND IMPLICATION IN HUMAN NAD BIOSYNTHESIS
J.Biol.Chem., 278, 2003
|
|
7VPX
 
 | | The cryo-EM structure of the human pre-A complex | | Descriptor: | 5SS, DnaJ homolog subfamily C member 8, PHD finger-like domain-containing protein 5A, ... | | Authors: | Zhang, X, Zhan, X, Shi, Y. | | Deposit date: | 2021-10-18 | | Release date: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanism for Branch Site Recognition and Proofreading During Prespliceosome Assembly
To Be Published
|
|
1NUP
 
 | | CRYSTAL STRUCTURE OF HUMAN CYTOSOLIC NMN/NaMN ADENYLYLTRANSFERASE COMPLEX WITH NMN | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, FKSG76, SULFATE ION | | Authors: | Zhang, X, Kurnasov, O.V, Karthikeyan, S, Grishin, N.V, Osterman, A.L, Zhang, H. | | Deposit date: | 2003-02-01 | | Release date: | 2003-06-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | STRUCTURAL CHARACTERIZATION OF A HUMAN CYTOSOLIC NMN/NaMN
ADENYLYLTRANSFERASE AND IMPLICATION IN HUMAN NAD BIOSYNTHESIS
J.Biol.Chem., 278, 2003
|
|
3AH5
 
 | | Crystal Structure of flavin dependent thymidylate synthase ThyX from helicobacter pylori complexed with FAD and dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Zhang, X, Zhang, J, Hu, Y, Zou, Q, Wang, D. | | Deposit date: | 2010-04-14 | | Release date: | 2011-04-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure and functional analysis of a flavin dependent thymidylate synthase from helicobacter pylori
To be Published
|
|
1NQV
 
 | | Crystal Structure of Lumazine Synthase from Aquifex aeolicus in Complex with Inhibitor: 5-nitroso-6-ribityl-amino-2,4(1H,3H)pyrimidinedione | | Descriptor: | 5-NITROSO-6-RIBITYL-AMINO-2,4(1H,3H)-PYRIMIDINEDIONE, 6,7-dimethyl-8-ribityllumazine synthase, PHOSPHATE ION | | Authors: | Zhang, X, Meining, W, Cushman, M, Haase, I, Fischer, M, Bacher, A, Ladenstein, R. | | Deposit date: | 2003-01-23 | | Release date: | 2004-01-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A structure-based model of the reaction catalyzed by lumazine synthase from Aquifex aeolicus.
J.Mol.Biol., 328, 2003
|
|
1NQW
 
 | | Crystal Structure of Lumazine Synthase from Aquifex aeolicus in Complex with Inhibitor: 5-(6-D-ribitylamino-2,4(1H,3H)pyrimidinedione-5-yl)-1-pentyl-phosphonic acid | | Descriptor: | 5-(6-D-RIBITYLAMINO-2,4(1H,3H)PYRIMIDINEDIONE-5-YL) PENTYL-1-PHOSPHONIC ACID, 6,7-dimethyl-8-ribityllumazine synthase | | Authors: | Zhang, X, Meining, W, Cushman, M, Haase, I, Fischer, M, Bacher, A, Ladenstein, R. | | Deposit date: | 2003-01-23 | | Release date: | 2004-01-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A structure-based model of the reaction catalyzed by lumazine synthase from Aquifex aeolicus.
J.Mol.Biol., 328, 2003
|
|
1NQX
 
 | | Crystal Structure of Lumazine Synthase from Aquifex aeolicus in Complex with Inhibitor: 3-(7-hydroxy-8-ribityllumazine-6-yl)propionic acid | | Descriptor: | 3-(7-HYDROXY-8-RIBITYLLUMAZINE-6-YL) PROPIONIC ACID, 6,7-dimethyl-8-ribityllumazine synthase, PHOSPHATE ION | | Authors: | Zhang, X, Meining, W, Cushman, M, Haase, I, Fischer, M, Bacher, A, Ladenstein, R. | | Deposit date: | 2003-01-23 | | Release date: | 2004-01-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | A structure-based model of the reaction catalyzed by lumazine synthase from Aquifex aeolicus.
J.Mol.Biol., 328, 2003
|
|
1NQU
 
 | | Crystal Structure of Lumazine Synthase from Aquifex aeolicus in Complex with Inhibitor: 6,7-dioxo-5H-8-ribitylaminolumazine | | Descriptor: | 6,7-DIOXO-5H-8-RIBITYLAMINOLUMAZINE, 6,7-dimethyl-8-ribityllumazine synthase, PHOSPHATE ION | | Authors: | Zhang, X, Meining, W, Cushman, M, Haase, I, Fischer, M, Bacher, A, Ladenstein, R. | | Deposit date: | 2003-01-23 | | Release date: | 2004-01-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A structure-based model of the reaction catalyzed by lumazine synthase from Aquifex aeolicus.
J.Mol.Biol., 328, 2003
|
|
8H0Z
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Locked-122 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-11-09 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
2RFD
 
 | | Crystal structure of the complex between the EGFR kinase domain and a Mig6 peptide | | Descriptor: | ERBB receptor feedback inhibitor 1, Epidermal growth factor receptor, SULFATE ION | | Authors: | Zhang, X, Pickin, K.A, Bose, R, Jura, N, Cole, P.A, Kuriyan, J. | | Deposit date: | 2007-09-28 | | Release date: | 2007-12-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Inhibition of the EGF receptor by binding of MIG6 to an activating kinase domain interface.
Nature, 450, 2007
|
|
2RF9
 
 | | Crystal structure of the complex between the EGFR kinase domain and a Mig6 peptide | | Descriptor: | ERBB receptor feedback inhibitor 1, Epidermal growth factor receptor | | Authors: | Zhang, X, Pickin, K.A, Bose, R, Jura, N, Cole, P.A, Kuriyan, J. | | Deposit date: | 2007-09-28 | | Release date: | 2007-12-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Inhibition of the EGF receptor by binding of MIG6 to an activating kinase domain interface.
Nature, 450, 2007
|
|
2RFE
 
 | | Crystal structure of the complex between the EGFR kinase domain and a Mig6 peptide | | Descriptor: | ERBB receptor feedback inhibitor 1, Epidermal growth factor receptor | | Authors: | Zhang, X, Pickin, K.A, Bose, R, Jura, N, Cole, P.A, Kuriyan, J. | | Deposit date: | 2007-09-28 | | Release date: | 2007-12-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Inhibition of the EGF receptor by binding of MIG6 to an activating kinase domain interface.
Nature, 450, 2007
|
|
2Y3A
 
 | | Crystal structure of p110beta in complex with icSH2 of p85beta and the drug GDC-0941 | | Descriptor: | 2-(1H-indazol-4-yl)-6-{[4-(methylsulfonyl)piperazin-1-yl]methyl}-4-morpholin-4-yl-thieno[3,2-d]pyrimidine, PHOSPHATIDYLINOSITOL 3-KINASE REGULATORY SUBUNIT BETA, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT BETA ISOFORM | | Authors: | Zhang, X, Vadas, O, Perisic, O, Williams, R.L. | | Deposit date: | 2010-12-20 | | Release date: | 2011-03-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of Lipid Kinase P110Beta-P85Beta Elucidates an Unusual Sh2-Domain-Mediated Inhibitory Mechanism.
Mol.Cell, 41, 2011
|
|
3OJI
 
 | | X-ray crystal structure of the Py13 -pyrabactin complex | | Descriptor: | 4-bromo-N-(pyridin-2-ylmethyl)naphthalene-1-sulfonamide, Abscisic acid receptor PYL3, SULFATE ION | | Authors: | Zhang, X, Zhang, Q, Wang, G, Chen, Z. | | Deposit date: | 2010-08-23 | | Release date: | 2011-08-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Complex Structures of the Abscisic Acid Receptor PYL3/RCAR13 Reveal a Unique Regulatory Mechanism
Structure, 20, 2012
|
|
8H10
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Locked-2 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-10-19 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H13
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x2 Disulfide (G400C and V969C), Closed Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-10-19 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H14
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x3 Disulfide (D414C and V969C), Locked-1 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, Spike glycoprotein | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-10-19 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H16
 
 | | Structure of SARS-CoV-1 Spike Protein (S/native) at pH 5.5, Open Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-11-09 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.35534 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H11
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Closed Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-11-09 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H12
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x2 Disulfide (G400C and V969C), Locked-2 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-11-09 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.44681 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H0Y
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Locked-112 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, LINOLEIC ACID, ... | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-11-09 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
