2H71
 
 | |
2H73
 
 | |
2H6X
 
 | |
2H6Y
 
 | |
2H74
 
 | |
2H75
 
 | |
2H76
 
 | |
2H6Z
 
 | | Crystal Structure of Thioredoxin Mutant E44D in Hexagonal (p61) Space Group | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Thioredoxin | | Authors: | Gavira, J.A, Godoy-Ruiz, R, Ibarra-Molero, B, Sanchez-Ruiz, J.M. | | Deposit date: | 2006-06-01 | | Release date: | 2007-05-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A stability pattern of protein hydrophobic mutations that reflects evolutionary structural optimization.
Biophys.J., 89, 2005
|
|
2H70
 
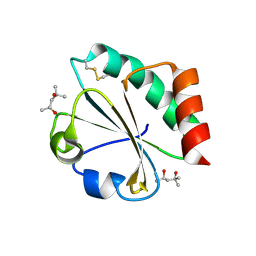 | | Crystal Structure of Thioredoxin Mutant D9E in Hexagonal (p61) Space Group | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Thioredoxin | | Authors: | Gavira, J.A, Godoy-Ruiz, R, Ibarra-Molero, B, Sanchez-Ruiz, J.M. | | Deposit date: | 2006-06-01 | | Release date: | 2007-05-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A stability pattern of protein hydrophobic mutations that reflects evolutionary structural optimization.
Biophys.J., 89, 2005
|
|
2H72
 
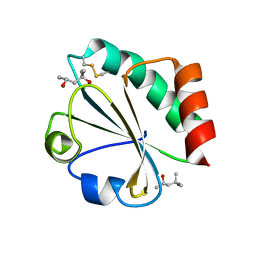 | |
5K8Q
 
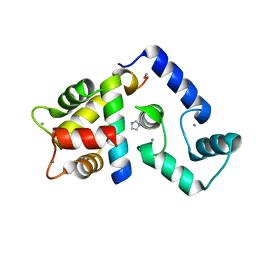 | | Crystal Structure of Calcium-loaded Calmodulin in complex with STRA6 CaMBP2-site peptide. | | Descriptor: | CALCIUM ION, Calmodulin, IMIDAZOLE, ... | | Authors: | Stowe, S.D, Clarke, O.B, Cavalier, M.C, Godoy-Ruiz, R, Mancia, F, Weber, D.J. | | Deposit date: | 2016-05-30 | | Release date: | 2016-08-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.739 Å) | | Cite: | Structure of the STRA6 receptor for retinol uptake.
Science, 353, 2016
|
|
4ULX
 
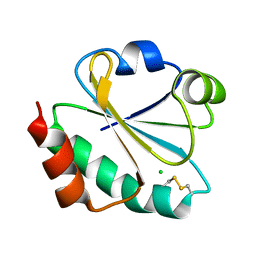 | | Crystal structure of ancestral thioredoxin, relative to the last common ancestor of the Cyanobacterial, Deinococcus and Thermus groups, LPBCA-L89K mutant. | | Descriptor: | CHLORIDE ION, LPBCA-L89K THIOREDOXIN | | Authors: | Gavira, J.A, Risso, V.A, Ibarra-Molero, B, Sanchez-Ruiz, J.M. | | Deposit date: | 2014-05-14 | | Release date: | 2014-11-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Mutational Studies on Resurrected Ancestral Proteins Reveal Conservation of Site-Specific Amino Acid Preferences Throughout Evolutionary History.
Mol.Biol.Evol., 32, 2015
|
|
5SY1
 
 | | Structure of the STRA6 receptor for retinol uptake in complex with calmodulin | | Descriptor: | CALCIUM ION, CHOLESTEROL, Calmodulin, ... | | Authors: | Clarke, O.B, Chen, Y, Mancia, F. | | Deposit date: | 2016-08-10 | | Release date: | 2016-08-24 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the STRA6 receptor for retinol uptake.
Science, 353, 2016
|
|
4FQO
 
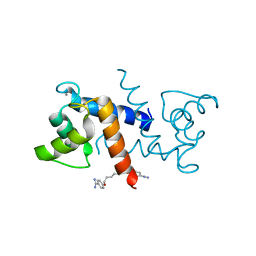 | | Crystal Structure of Calcium-Loaded S100B Bound to SBi4211 | | Descriptor: | 4,4'-[heptane-1,7-diylbis(oxy)]dibenzenecarboximidamide, CALCIUM ION, Protein S100-B | | Authors: | McKnight, L.E, Raman, E.P, Bezawada, P, Kudrimoti, S, Wilder, P.T, Hartman, K.G, Toth, E.A, Coop, A, MacKerrell, A.D, Weber, D.J. | | Deposit date: | 2012-06-25 | | Release date: | 2012-10-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-Based Discovery of a Novel Pentamidine-Related Inhibitor of the Calcium-Binding Protein S100B.
ACS Med Chem Lett, 3, 2012
|
|
6UAI
 
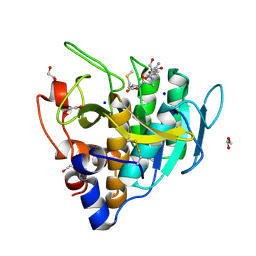 | | Imidazole-triggered RAS-specific subtilisin SUBT_BACAM complexed with YSAM peptide | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Toth, E.A, Bryan, P.N, Orban, J. | | Deposit date: | 2019-09-10 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Engineering subtilisin proteases that specifically degrade active RAS.
Commun Biol, 4, 2021
|
|
6UAO
 
 | | Imidazole-triggered RAS-specific subtilisin SUBT_BACAM complexed with the peptide EEYSAM | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Peptide EEYSAM, ... | | Authors: | Toth, E.A, Bryan, P.N, Orban, J. | | Deposit date: | 2019-09-11 | | Release date: | 2020-09-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Engineering subtilisin proteases that specifically degrade active RAS.
Commun Biol, 4, 2021
|
|
6UBE
 
 | | Azide-triggered subtilisin SUBT_BACAM complexed with the peptide LFRAL | | Descriptor: | AZIDE ION, GLYCEROL, Peptide LFRAL, ... | | Authors: | Toth, E.A, Bryan, P.N, Orban, J, Gallagher, D.T, Custer, G. | | Deposit date: | 2019-09-11 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Engineering subtilisin proteases that specifically degrade active RAS.
Commun Biol, 4, 2021
|
|
6U9L
 
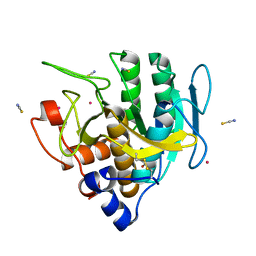 | | Imidazole-triggered RAS-specific subtilisin SUBT_BACAM | | Descriptor: | GLYCEROL, POTASSIUM ION, SUBTILISIN BPN', ... | | Authors: | Toth, E.A, Bryan, P.N, Orban, J. | | Deposit date: | 2019-09-09 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Engineering subtilisin proteases that specifically degrade active RAS.
Commun Biol, 4, 2021
|
|
6UWO
 
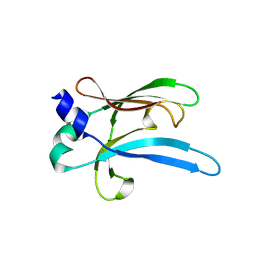 | |
6UWT
 
 | |
6UWR
 
 | |
6UWI
 
 | | Crystal structure of the Clostridium difficile translocase CDTb | | Descriptor: | ADP-ribosyltransferase binding component, CALCIUM ION | | Authors: | Pozharski, E. | | Deposit date: | 2019-11-05 | | Release date: | 2020-01-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structure of the cell-binding component of theClostridium difficilebinary toxin reveals a di-heptamer macromolecular assembly.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
