1H1B
 
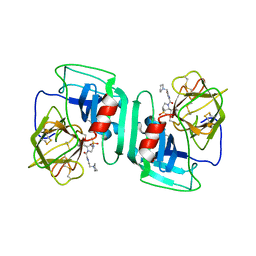 | | Crystal structure of human neutrophil elastase complexed with an inhibitor (GW475151) | | Descriptor: | (2S)-3-METHYL-2-((2R,3S)-3-[(METHYLSULFONYL)AMINO]-1-{[2-(PYRROLIDIN-1-YLMETHYL)-1,3-OXAZOL-4-YL]CARBONYL}PYRROLIDIN-2-YL)BUTANOIC ACID, LEUKOCYTE ELASTASE, alpha-L-fucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Macdonald, S.J.F, Dowle, M.D, Harrison, L.A, Clarke, G.D.E, Inglis, G.G.A, Johnson, M.R, Smith, R.A, Amour, A, Fleetwood, G, Humphreys, D.C, Molloy, C.R, Dixon, M, Godward, R.E, Wonacott, A.J, Singh, O.M.P, Hodgson, S.T, Hardy, G.W. | | Deposit date: | 2002-07-05 | | Release date: | 2002-08-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Further Pyrrolidine Trans-Lactams as Inhibitors of Human Neutrophil Elastase (Hne) with Potential as Development Candidates and the Crystal Structure of Hne Complexed with an Inhibitor (Gw475151)
J.Med.Chem., 45, 2002
|
|
1KQQ
 
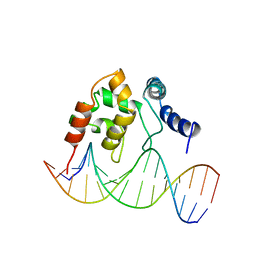 | | Solution Structure of the Dead ringer ARID-DNA Complex | | Descriptor: | 5'-D(*CP*CP*AP*CP*AP*TP*CP*AP*AP*TP*AP*CP*AP*GP*G)-3', 5'-D(*CP*CP*TP*GP*TP*AP*TP*TP*GP*AP*TP*GP*TP*GP*G)-3', DEAD RINGER PROTEIN | | Authors: | Iwahara, J, Iwahara, M, Daughdrill, G.W, Ford, J, Clubb, R.T. | | Deposit date: | 2002-01-07 | | Release date: | 2002-03-06 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | The structure of the Dead ringer-DNA complex reveals how AT-rich interaction domains (ARIDs) recognize DNA.
EMBO J., 21, 2002
|
|
170D
 
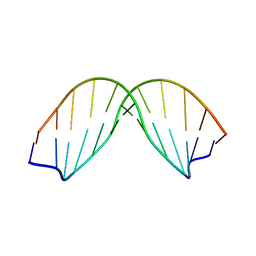 | | SOLUTION STRUCTURE OF A DNA DODECAMER CONTAINING THE ANTI-NEOPLASTIC AGENT ARABINOSYLCYTOSINE: COMBINED USE OF NMR, RESTRAINED MOLECULAR DYNAMICS AND FULL RELAXATION MATRIX REFINEMENT | | Descriptor: | DNA/RNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*T)-R(P*CAR)-D(P*GP*CP*G)-3') | | Authors: | Schweitzer, B.I, Mikita, T, Kellogg, G.W, Gardner, K.H, Beardsley, G.P. | | Deposit date: | 1994-03-14 | | Release date: | 1994-07-31 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a DNA dodecamer containing the anti-neoplastic agent arabinosylcytosine: combined use of NMR, restrained molecular dynamics, and full relaxation matrix refinement.
Biochemistry, 33, 1994
|
|
7XK2
 
 | | Cryo-EM Structure of Human Niacin Receptor HCA2-Gi protein complex | | Descriptor: | 2-[[2,2-dimethyl-3-[3-(5-oxidanylpyridin-2-yl)-1,2,4-oxadiazol-5-yl]propanoyl]amino]cyclohexene-1-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yang, Y, Kang, H.J, Gao, R.G, Wang, J.J, Han, G.W, DiBerto, J.F, Wu, L.J, Tong, J.H, Qu, L, Wu, Y.R, Pileski, R, Li, X.M, Zhang, X.C, Zhao, S.W, Kenakin, T, Wang, Q, Stevens, R.C, Peng, W, Roth, B.L, Rao, Z.H, Liu, Z.J. | | Deposit date: | 2022-04-19 | | Release date: | 2023-02-22 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the human niacin receptor HCA2-G i signalling complex.
Nat Commun, 14, 2023
|
|
1ALE
 
 | |
1ALF
 
 | |
171D
 
 | | SOLUTION STRUCTURE OF A DNA DODECAMER CONTAINING THE ANTI-NEOPLASTIC AGENT ARABINOSYLCYTOSINE: COMBINED USE OF NMR, RESTRAINED MOLECULAR DYNAMICS AND FULL RELAXATION MATRIX REFINEMENT | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Schweitzer, B.I, Mikita, T, Kellogg, G.W, Gardner, K.H, Beardsley, G.P. | | Deposit date: | 1994-03-14 | | Release date: | 1994-07-31 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a DNA dodecamer containing the anti-neoplastic agent arabinosylcytosine: combined use of NMR, restrained molecular dynamics, and full relaxation matrix refinement.
Biochemistry, 33, 1994
|
|
1B8X
 
 | | GLUTATHIONE S-TRANSFERASE FUSED WITH THE NUCLEAR MATRIX TARGETING SIGNAL OF THE TRANSCRIPTION FACTOR AML-1 | | Descriptor: | PROTEIN (AML-1B) | | Authors: | Tang, L, Guo, B, Van Wijnen, A.J, Lian, J.B, Stein, J.L, Stein, G.S, Zhou, G.W. | | Deposit date: | 1999-02-03 | | Release date: | 1999-04-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Preliminary crystallographic study of glutathione S-transferase fused with the nuclear matrix targeting signal of the transcription factor AML-1/CBF-alpha2.
J.Struct.Biol., 123, 1998
|
|
7ZL9
 
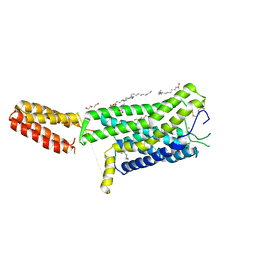 | | Crystal structure of human GPCR Niacin receptor (HCA2) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, DI(HYDROXYETHYL)ETHER, Hydroxycarboxylic acid receptor 2,Soluble cytochrome b562, ... | | Authors: | Yang, Y, Kang, H.J, Gao, R.G, Wang, J.J, Han, G.W, FiBerto, J.F, Wu, L.J, Tong, J.H, Qu, L, Wu, Y.R, Pileski, R, Li, X.M, Zhang, X.C, Zhao, S.W, Kenakin, T, Wang, Q, Stevens, R.C, Peng, W, Roth, B.L, Rao, Z.H, Liu, Z.J. | | Deposit date: | 2022-04-14 | | Release date: | 2023-04-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into the human niacin receptor HCA2-G i signalling complex.
Nat Commun, 14, 2023
|
|
7ZLY
 
 | | Crystal structure of human GPCR Niacin receptor (HCA2) expressed from Spodoptera frugiperda | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Hydroxycarboxylic acid receptor 2,Soluble cytochrome b562, OLEIC ACID | | Authors: | Yang, Y, Kang, H.J, Gao, R.G, Wang, J.J, FiBerto, J.F, Wu, L.J, Tong, J.H, Han, G.W, Qu, L, Wu, Y.R, Pileski, R, Li, X.M, Zhang, X.C, Zhao, S.W, Kenakin, T, Wang, Q, Stevens, R.C, Peng, W, Roth, B.L, Rao, Z.H, Liu, Z.J. | | Deposit date: | 2022-04-17 | | Release date: | 2023-04-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into the human niacin receptor HCA2-G i signalling complex.
Nat Commun, 14, 2023
|
|
7ZWO
 
 | | Crystal structure of human BCL6 BTB domain in complex with compound 2 | | Descriptor: | (5~{S},7~{R})-5-(4-chlorophenyl)-7-(2,3,4-trimethoxyphenyl)-4,5,6,7-tetrahydro-[1,2,4]triazolo[1,5-a]pyrimidine, 1,2-ETHANEDIOL, ALA-TRP-VAL-ILE-PRO-ALA, ... | | Authors: | Collie, G.W, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2022-05-19 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Discovering cell-active BCL6 inhibitors: effectively combining biochemical HTS with multiple biophysical techniques, X-ray crystallography and cell-based assays.
Sci Rep, 12, 2022
|
|
7ZWS
 
 | | Crystal structure of human BCL6 BTB domain in complex with compound 13 | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-cyano-6-thiophen-2-yl-4-(trifluoromethyl)pyridin-2-yl]sulfanyl-2-phenyl-ethanoic acid, ALA-TRP-VAL-ILE-PRO-ALA, ... | | Authors: | Collie, G.W, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2022-05-19 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Discovering cell-active BCL6 inhibitors: effectively combining biochemical HTS with multiple biophysical techniques, X-ray crystallography and cell-based assays.
Sci Rep, 12, 2022
|
|
7ZWR
 
 | | Crystal structure of human BCL6 BTB domain in complex with compound 11 | | Descriptor: | 1,2-ETHANEDIOL, 2-[2-[[(6~{S})-3-cyano-6-methyl-4-(trifluoromethyl)-5,6,7,8-tetrahydroquinolin-2-yl]sulfanyl]ethanoylamino]ethanoic acid, ALA-TRP-VAL-ILE-PRO-ALA, ... | | Authors: | Collie, G.W, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2022-05-19 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Discovering cell-active BCL6 inhibitors: effectively combining biochemical HTS with multiple biophysical techniques, X-ray crystallography and cell-based assays.
Sci Rep, 12, 2022
|
|
7ZWX
 
 | | Crystal structure of human BCL6 BTB domain in complex with compound 19 | | Descriptor: | 1,2-ETHANEDIOL, 6-[1,3-benzodioxol-5-ylmethyl(methyl)amino]-1-~{tert}-butyl-5~{H}-pyrazolo[3,4-d]pyrimidin-4-one, ALA-TRP-VAL-ILE-PRO-ALA, ... | | Authors: | Collie, G.W, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2022-05-19 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Discovering cell-active BCL6 inhibitors: effectively combining biochemical HTS with multiple biophysical techniques, X-ray crystallography and cell-based assays.
Sci Rep, 12, 2022
|
|
7ZWZ
 
 | | Crystal structure of human BCL6 BTB domain in complex with compound 22 | | Descriptor: | 1,2-ETHANEDIOL, ALA-TRP-VAL-ILE-PRO-ALA, B-cell lymphoma 6 protein, ... | | Authors: | Collie, G.W, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2022-05-19 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Discovering cell-active BCL6 inhibitors: effectively combining biochemical HTS with multiple biophysical techniques, X-ray crystallography and cell-based assays.
Sci Rep, 12, 2022
|
|
7ZWU
 
 | | Crystal structure of human BCL6 BTB domain in complex with compound 15 | | Descriptor: | 1,2-ETHANEDIOL, ALA-TRP-VAL-ILE-PRO-ALA, B-cell lymphoma 6 protein, ... | | Authors: | Collie, G.W, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2022-05-19 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Discovering cell-active BCL6 inhibitors: effectively combining biochemical HTS with multiple biophysical techniques, X-ray crystallography and cell-based assays.
Sci Rep, 12, 2022
|
|
7ZWV
 
 | | Crystal structure of human BCL6 BTB domain in complex with compound 17 | | Descriptor: | 1,2-ETHANEDIOL, 1,3-dimethyl-5-[[6-(phenylmethylsulfanyl)pyrimidin-4-yl]amino]benzimidazol-2-one, ALA-TRP-VAL-ILE-PRO-ALA, ... | | Authors: | Collie, G.W, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2022-05-19 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Discovering cell-active BCL6 inhibitors: effectively combining biochemical HTS with multiple biophysical techniques, X-ray crystallography and cell-based assays.
Sci Rep, 12, 2022
|
|
8AN8
 
 | | Crystal structure of wild-type c-MET bound by compound 7. | | Descriptor: | 3-[bis(fluoranyl)methyl]-~{N}-methyl-~{N}-[(1~{R})-8-methyl-5-(3-methyl-1~{H}-indazol-6-yl)-1,2,3,4-tetrahydronaphthalen-1-yl]pyridine-2-carboxamide, Hepatocyte growth factor receptor, SULFATE ION | | Authors: | Collie, G.W. | | Deposit date: | 2022-08-04 | | Release date: | 2022-08-31 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.394 Å) | | Cite: | Discovery of a selective c-MET inhibitor with a novel binding mode.
Bioorg.Med.Chem.Lett., 75, 2022
|
|
8ANS
 
 | | Crystal structure of D1228V c-MET bound by compound 1. | | Descriptor: | 3-[bis(fluoranyl)methyl]-~{N}-methyl-~{N}-[(1~{R})-8-methyl-5-(3-methyl-1~{H}-indazol-6-yl)-1,2,3,4-tetrahydronaphthalen-1-yl]pyridine-2-carboxamide, GLYCEROL, Hepatocyte growth factor receptor | | Authors: | Collie, G.W. | | Deposit date: | 2022-08-05 | | Release date: | 2022-08-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Discovery of a selective c-MET inhibitor with a novel binding mode.
Bioorg.Med.Chem.Lett., 75, 2022
|
|
4EIY
 
 | | Crystal structure of the chimeric protein of A2aAR-BRIL in complex with ZM241385 at 1.8A resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | Authors: | Liu, W, Chun, E, Thompson, A.A, Chubukov, P, Xu, F, Katritch, V, Han, G.W, Heitman, L.H, Ijzerman, A.P, Cherezov, V, Stevens, R.C, GPCR Network (GPCR) | | Deposit date: | 2012-04-06 | | Release date: | 2012-07-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for allosteric regulation of GPCRs by sodium ions.
Science, 337, 2012
|
|
5HBT
 
 | | Complex structure of Fab35 and human nAChR alpha1 | | Descriptor: | Acetylcholine receptor subunit alpha 1, Alpha-bungarotoxin isoform V31, Fab35, ... | | Authors: | Noridomi, K, Watanabe, G, Hansen, M.N, Han, G.W, Chen, L. | | Deposit date: | 2016-01-02 | | Release date: | 2017-05-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural insights into the molecular mechanisms of myasthenia gravis and their therapeutic implications.
Elife, 6, 2017
|
|
5JGK
 
 | | Crystal structure of GtmA in complex with SAH | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, UbiE/COQ5 family methyltransferase, ... | | Authors: | Dolan, S.K, Bock, T, Hering, V, Jones, G.W, Blankenfeldt, W, Dolye, S. | | Deposit date: | 2016-04-20 | | Release date: | 2017-03-01 | | Last modified: | 2018-03-28 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Structural, mechanistic and functional insight into gliotoxinbis-thiomethylation inAspergillus fumigatus.
Open Biol, 7, 2017
|
|
2GPN
 
 | | 100 K STRUCTURE OF GLYCOGEN PHOSPHORYLASE AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | GLYCOGEN PHOSPHORYLASE B | | Authors: | Gregoriou, M, Noble, M.E.M, Watson, K.A, Garman, E.F, Krulle, T.M, De La Fuente, C, Fleet, G.W.J, Oikonomakos, N.G, Johnson, L.N. | | Deposit date: | 1998-03-26 | | Release date: | 1998-07-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The structure of a glycogen phosphorylase glucopyranose spirohydantoin complex at 1.8 A resolution and 100 K: the role of the water structure and its contribution to binding.
Protein Sci., 7, 1998
|
|
5JGJ
 
 | | Crystal structure of GtmA | | Descriptor: | UbiE/COQ5 family methyltransferase, putative | | Authors: | Dolan, S.K, Bock, T, Hering, V, Jones, G.W, Blankenfeldt, W, Doyle, S. | | Deposit date: | 2016-04-20 | | Release date: | 2017-03-01 | | Last modified: | 2018-03-28 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural, mechanistic and functional insight into gliotoxinbis-thiomethylation inAspergillus fumigatus.
Open Biol, 7, 2017
|
|
5JGL
 
 | | Crystal structure of GtmA in complex with S-Adenosylmethionine | | Descriptor: | S-ADENOSYLMETHIONINE, SODIUM ION, UbiE/COQ5 family methyltransferase, ... | | Authors: | Dolan, S.K, Bock, T, Hering, V, Jones, G.W, Blankenfeldt, W, Doyle, S. | | Deposit date: | 2016-04-20 | | Release date: | 2017-03-01 | | Last modified: | 2018-03-28 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural, mechanistic and functional insight into gliotoxinbis-thiomethylation inAspergillus fumigatus.
Open Biol, 7, 2017
|
|
