4Z36
 
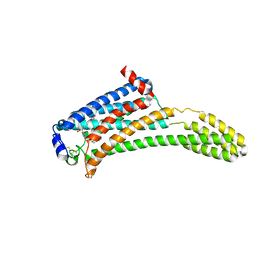 | | Crystal Structure of Human Lysophosphatidic Acid Receptor 1 in complex with ONO-3080573 | | Descriptor: | (2S)-2,3-dihydroxypropyl (7Z)-tetradec-7-enoate, 1-(4-{[(2S,3R)-2-(2,3-dihydro-1H-inden-2-yloxy)-3-(3,5-dimethoxy-4-methylphenyl)-3-hydroxypropyl]oxy}phenyl)cyclopropanecarboxylic acid, Lysophosphatidic acid receptor 1,Soluble cytochrome b562 | | Authors: | Chrencik, J.E, Roth, C.B, Terakado, M, Kurata, H, Omi, R, Kihara, Y, Warshaviak, D, Nakade, S, Asmar-Rovira, G, Mileni, M, Mizuno, H, Griffith, M.T, Rodgers, C, Han, G.W, Velasquez, J, Chun, J, Stevens, R.C, Hanson, M.A, GPCR Network (GPCR) | | Deposit date: | 2015-03-30 | | Release date: | 2015-06-03 | | Last modified: | 2015-07-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of Antagonist Bound Human Lysophosphatidic Acid Receptor 1.
Cell, 161, 2015
|
|
1PJG
 
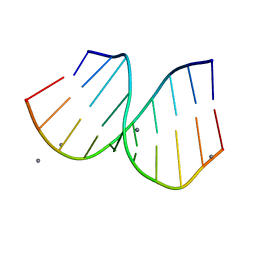 | | RNA/DNA Hybrid Decamer of CAAAGAAAAG/CTTTTCTTTG | | Descriptor: | 5'-D(*CP*TP*TP*TP*TP*CP*TP*TP*TP*G)-3', 5'-R(*CP*AP*AP*AP*GP*AP*AP*AP*AP*G)-3', CALCIUM ION | | Authors: | Kopka, M.L, Lavelle, L, Han, G.W, Ng, H.-L, Dickerson, R.E. | | Deposit date: | 2003-06-02 | | Release date: | 2003-12-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | An unusual sugar conformation in the structure of an RNA/DNA decamer of the polypurine tract may affect recognition by RNase H.
J.Mol.Biol., 334, 2003
|
|
1PJO
 
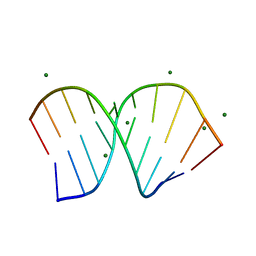 | | Crystal Structure of an RNA/DNA hybrid of HIV-1 PPT | | Descriptor: | 5'-D(*CP*TP*TP*TP*TP*CP*TP*TP*TP*G)-3', 5'-R(*CP*AP*AP*AP*GP*AP*AP*AP*AP*G)-3', MAGNESIUM ION | | Authors: | Kopka, M.L, Lavelle, L, Han, G.W, Ng, H.-L, Dickerson, R.E. | | Deposit date: | 2003-06-03 | | Release date: | 2003-12-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | An Unusual Sugar Conformation in the Structure of an RNA/DNA
Decamer of the Polypurine Tract May Affect Recognition by RNase H
J.Mol.Biol., 334, 2003
|
|
4ZUD
 
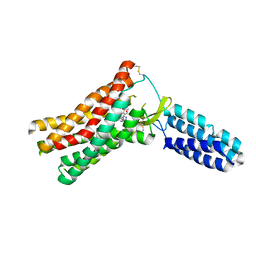 | | Crystal Structure of Human Angiotensin Receptor in Complex with Inverse Agonist Olmesartan at 2.8A resolution. | | Descriptor: | Chimera protein of Soluble cytochrome b562 and Type-1 angiotensin II receptor, Olmesartan | | Authors: | Zhang, H, Unal, H, Desnoyer, R, Han, G.W, Patel, N, Katritch, V, Karnik, S.S, Cherezov, V, Stevens, R.C, GPCR Network (GPCR) | | Deposit date: | 2015-05-15 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for Ligand Recognition and Functional Selectivity at Angiotensin Receptor.
J.Biol.Chem., 290, 2015
|
|
4ZWJ
 
 | | Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser | | Descriptor: | Chimera protein of human Rhodopsin, mouse S-arrestin, and T4 Endolysin | | Authors: | Kang, Y, Zhou, X.E, Gao, X, He, Y, Liu, W, Ishchenko, A, Barty, A, White, T.A, Yefanov, O, Han, G.W, Xu, Q, de Waal, P.W, Ke, J, Tan, M.H.E, Zhang, C, Moeller, A, West, G.M, Pascal, B, Eps, N.V, Caro, L.N, Vishnivetskiy, S.A, Lee, R.J, Suino-Powell, K.M, Gu, X, Pal, K, Ma, J, Zhi, X, Boutet, S, Williams, G.J, Messerschmidt, M, Gati, C, Zatsepin, N.A, Wang, D, James, D, Basu, S, Roy-Chowdhury, S, Conrad, C, Coe, J, Liu, H, Lisova, S, Kupitz, C, Grotjohann, I, Fromme, R, Jiang, Y, Tan, M, Yang, H, Li, J, Wang, M, Zheng, Z, Li, D, Howe, N, Zhao, Y, Standfuss, J, Diederichs, K, Dong, Y, Potter, C.S, Carragher, B, Caffrey, M, Jiang, H, Chapman, H.N, Spence, J.C.H, Fromme, P, Weierstall, U, Ernst, O.P, Katritch, V, Gurevich, V.V, Griffin, P.R, Hubbell, W.L, Stevens, R.C, Cherezov, V, Melcher, K, Xu, H.E, GPCR Network (GPCR) | | Deposit date: | 2015-05-19 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.302 Å) | | Cite: | Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser.
Nature, 523, 2015
|
|
6D0T
 
 | | De novo design of a fluorescence-activating beta barrel - BB1 | | Descriptor: | BB1 | | Authors: | Dou, J, Vorobieva, A.A, Sheffler, W, Doyle, L.A, Park, H, Bick, M.J, Mao, B, Foight, G.W, Lee, M, Carter, L, Sankaran, B, Ovchinnikov, S, Marcos, E, Huang, P, Vaughan, J.C, Stoddard, B.L, Baker, D. | | Deposit date: | 2018-04-10 | | Release date: | 2018-09-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | De novo design of a fluorescence-activating beta-barrel.
Nature, 561, 2018
|
|
6DCG
 
 | | Discovery of MK-8353: An Orally Bioavailable Dual Mechanism ERK Inhibitor for Oncology | | Descriptor: | (3S)-3-(methylsulfanyl)-1-(2-{4-[4-(1-methyl-1H-1,2,4-triazol-3-yl)phenyl]-3,6-dihydropyridin-1(2H)-yl}-2-oxoethyl)-N-(3-{6-[(propan-2-yl)oxy]pyridin-3-yl}-1H-indazol-5-yl)pyrrolidine-3-carboxamide, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Boga, S.B, Deng, Y, Zhu, L, Nan, Y, Cooper, A, Shipps Jr, G.W, Doll, R, Shih, N, Zhu, H, Sun, R, Wang, T, Paliwal, S, Tsui, H, Gao, X, Yao, X, Desai, J, Wang, J, Alhassan, A.B, Kelly, J, Patel, M, Muppalla, K, Gudipati, S, Zhang, L, Buevich, A, Hesk, D, Carr, D, Dayananth, P, Mei, H, Cox, K, Sherborne, B, Hruza, A.W, Xiao, L, Jin, W, Long, B, Liu, G, Taylor, S.A, Kirschmeier, P, Windsor, W.T, Bishop, R, Samatar, A.A. | | Deposit date: | 2018-05-06 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | MK-8353: Discovery of an Orally Bioavailable Dual Mechanism ERK Inhibitor for Oncology.
ACS Med Chem Lett, 9, 2018
|
|
1B4D
 
 | | AMIDOCARBAMATE INHIBITOR OF GLYCOGEN PHOSPHORYLASE | | Descriptor: | 1-DEOXY-1-METHOXYCARBAMIDO-BETA-D-GLUCO-2-HEPTULOPYRANOSONAMIDE, INOSINIC ACID, PROTEIN (GLYCOGEN PHOSPHORYLASE B), ... | | Authors: | Tsitsanou, K.E, Oikonomakos, N.G, Zographos, S.E, Skamnaki, V.T, Gregoriou, M, Watson, K.A, Johnson, L.N, Fleet, G.W.J. | | Deposit date: | 1998-12-18 | | Release date: | 1998-12-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Effects of commonly used cryoprotectants on glycogen phosphorylase activity and structure.
Protein Sci., 8, 1999
|
|
3KSV
 
 | |
3KPA
 
 | |
1GWZ
 
 | | CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF THE PROTEIN TYROSINE PHOSPHATASE SHP-1 | | Descriptor: | SHP-1 | | Authors: | Yang, J, Liang, X, Niu, T, Meng, W, Zhao, Z, Zhou, G.W. | | Deposit date: | 1998-08-22 | | Release date: | 1999-08-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the catalytic domain of protein-tyrosine phosphatase SHP-1.
J.Biol.Chem., 273, 1998
|
|
1JVO
 
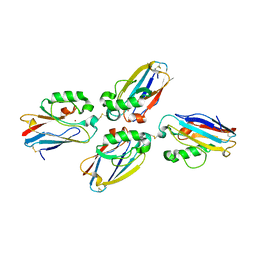 | | Azurin dimer, crosslinked via disulfide bridge | | Descriptor: | Azurin, COPPER (II) ION | | Authors: | van Amsterdam, I.M.C, Ubbink, M, Einsle, O, Messerschmidt, A, Merli, A, Cavazzini, D, Rossi, G.L, Canters, G.W. | | Deposit date: | 2001-08-30 | | Release date: | 2002-01-04 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Dramatic modulation of electron transfer in protein complexes by crosslinking
Nat.Struct.Biol., 9, 2002
|
|
1HKS
 
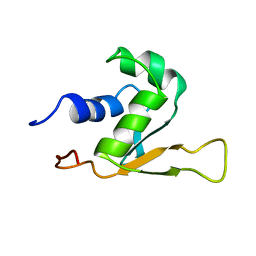 | | SOLUTION STRUCTURE OF THE DNA-BINDING DOMAIN OF DROSOPHILA HEAT SHOCK TRANSCRIPTION FACTOR | | Descriptor: | HEAT-SHOCK TRANSCRIPTION FACTOR | | Authors: | Vuister, G.W, Kim, S.-J, Orosz, A, Marquardt, J.L, Wu, C, Bax, A. | | Deposit date: | 1994-07-18 | | Release date: | 1994-09-30 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA-binding domain of Drosophila heat shock transcription factor.
Nat.Struct.Biol., 1, 1994
|
|
1HKT
 
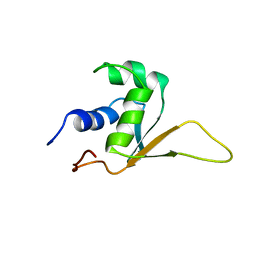 | | SOLUTION STRUCTURE OF THE DNA-BINDING DOMAIN OF DROSOPHILA HEAT SHOCK TRANSCRIPTION FACTOR | | Descriptor: | HEAT-SHOCK TRANSCRIPTION FACTOR | | Authors: | Vuister, G.W, Kim, S.-J, Orosz, A, Marquardt, J.L, Wu, C, Bax, A. | | Deposit date: | 1994-07-18 | | Release date: | 1994-09-30 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA-binding domain of Drosophila heat shock transcription factor.
Nat.Struct.Biol., 1, 1994
|
|
1KJV
 
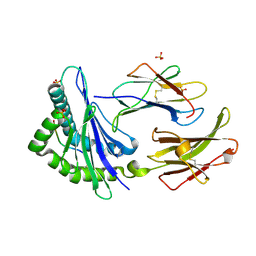 | | TAP-B-associated rat MHC class I molecule | | Descriptor: | Mature alpha chain of major histocompatibility complex class I antigen (HEAVY CHAIN), SULFATE ION, beta-2-microglobulin, ... | | Authors: | Rudolph, M.G, Stevens, J, Speir, J.A, Trowsdale, J, Butcher, G.W, Joly, E, Wilson, I.A. | | Deposit date: | 2001-12-05 | | Release date: | 2002-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal structures of two rat MHC class Ia (RT1-A) molecules that are associated differentially
with peptide transporter alleles TAP-A and TAP-B.
J.Mol.Biol., 324, 2002
|
|
1KJM
 
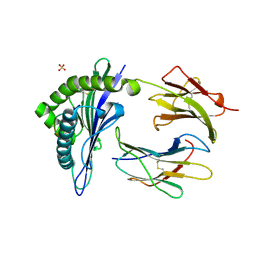 | | TAP-A-associated rat MHC class I molecule | | Descriptor: | B6 Peptide, RT1 class I histocompatibility antigen, AA alpha chain, ... | | Authors: | Rudolph, M.G, Stevens, J, Speir, J.A, Trowsdale, J, Butcher, G.W, Joly, E, Wilson, I.A. | | Deposit date: | 2001-12-04 | | Release date: | 2002-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structures of two rat MHC class Ia (RT1-A) molecules that are associated differentially
with peptide transporter alleles TAP-A and TAP-B.
J.Mol.Biol., 324, 2002
|
|
1JVL
 
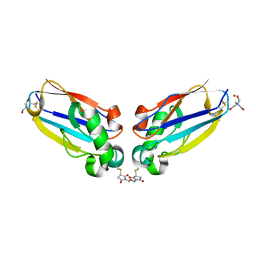 | | Azurin dimer, covalently crosslinked through bis-maleimidomethylether | | Descriptor: | 1-[PYRROL-1-YL-2,5-DIONE-METHOXYMETHYL]-PYRROLE-2,5-DIONE, Azurin, COPPER (II) ION, ... | | Authors: | van Amsterdam, I.M.C, Ubbink, M, Einsle, O, Messerschmidt, A, Merli, A, Cavazzini, D, Rossi, G.L, Canters, G.W. | | Deposit date: | 2001-08-30 | | Release date: | 2002-01-04 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dramatic modulation of electron transfer in protein complexes by crosslinking
Nat.Struct.Biol., 9, 2002
|
|
8OWG
 
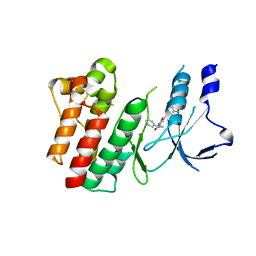 | | Crystal structure of D1228V c-MET bound by compound 2 | | Descriptor: | 5-[3,5-bis(fluoranyl)phenyl]-1-[(1S)-1-phenylethyl]pyrimidine-2,4-dione, Hepatocyte growth factor receptor | | Authors: | Collie, G.W. | | Deposit date: | 2023-04-27 | | Release date: | 2023-07-05 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (2.631 Å) | | Cite: | Discovery and Optimization of the First ATP Competitive Type-III c-MET Inhibitor.
J.Med.Chem., 66, 2023
|
|
8OUU
 
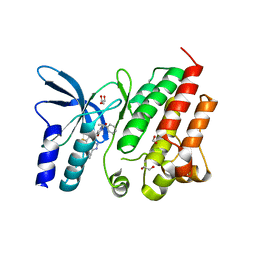 | | Crystal structure of D1228V c-MET bound by compound 29 | | Descriptor: | 1,2-ETHANEDIOL, 5-(3-ethynyl-5-fluoranyl-1H-indazol-7-yl)-1-[(1S)-1-phenylethyl]pyrimidine-2,4-dione, FORMIC ACID, ... | | Authors: | Collie, G.W. | | Deposit date: | 2023-04-24 | | Release date: | 2023-07-05 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Discovery and Optimization of the First ATP Competitive Type-III c-MET Inhibitor.
J.Med.Chem., 66, 2023
|
|
8OUV
 
 | | Crystal structure of D1228V c-MET bound by compound 15 | | Descriptor: | 5-(1H-indazol-7-yl)-1-[(1S)-1-phenylethyl]pyrimidine-2,4-dione, CHLORIDE ION, Hepatocyte growth factor receptor | | Authors: | Collie, G.W. | | Deposit date: | 2023-04-24 | | Release date: | 2023-07-05 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (1.783 Å) | | Cite: | Discovery and Optimization of the First ATP Competitive Type-III c-MET Inhibitor.
J.Med.Chem., 66, 2023
|
|
8OVZ
 
 | | Crystal structure of D1228V c-MET bound by compound 16 | | Descriptor: | 1-[(1S)-1-[3-(1H-imidazol-4-yl)phenyl]ethyl]-5-(1H-indazol-7-yl)pyrimidine-2,4-dione, Hepatocyte growth factor receptor, IODIDE ION | | Authors: | Collie, G.W. | | Deposit date: | 2023-04-26 | | Release date: | 2023-07-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Discovery and Optimization of the First ATP Competitive Type-III c-MET Inhibitor.
J.Med.Chem., 66, 2023
|
|
8OW3
 
 | | Crystal structure of wild-type c-MET bound by compound 2 | | Descriptor: | 5-[3,5-bis(fluoranyl)phenyl]-1-[(1S)-1-phenylethyl]pyrimidine-2,4-dione, Hepatocyte growth factor receptor | | Authors: | Collie, G.W. | | Deposit date: | 2023-04-26 | | Release date: | 2023-07-05 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Discovery and Optimization of the First ATP Competitive Type-III c-MET Inhibitor.
J.Med.Chem., 66, 2023
|
|
8OV7
 
 | | Crystal structure of D1228V c-MET bound by compound 10 | | Descriptor: | 5-[3,5-bis(fluoranyl)phenyl]-1-[(1S)-1-[3-(1H-imidazol-5-yl)phenyl]ethyl]pyrimidine-2,4-dione, Hepatocyte growth factor receptor | | Authors: | Collie, G.W. | | Deposit date: | 2023-04-25 | | Release date: | 2023-07-05 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery and Optimization of the First ATP Competitive Type-III c-MET Inhibitor.
J.Med.Chem., 66, 2023
|
|
1B64
 
 | | SOLUTION STRUCTURE OF THE GUANINE NUCLEOTIDE EXCHANGE FACTOR DOMAIN FROM HUMAN ELONGATION FACTOR-ONE BETA, NMR, 20 STRUCTURES | | Descriptor: | ELONGATION FACTOR 1-BETA | | Authors: | Perez, J.M.J, Siegal, G, Kriek, J, Hard, K, Dijk, J, Canters, G.W, Moller, W. | | Deposit date: | 1999-01-20 | | Release date: | 1999-05-18 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the guanine nucleotide exchange domain of human elongation factor 1beta reveals a striking resemblance to that of EF-Ts from Escherichia coli.
Structure Fold.Des., 7, 1999
|
|
1FS4
 
 | | Structures of glycogen phosphorylase-inhibitor complexes and the implications for structure-based drug design | | Descriptor: | 1-DEOXY-1-METHOXYCARBAMIDO-BETA-D-GLUCO-2-HEPTULOPYRANOSONAMIDE, GLYCOGEN PHOSPHORYLASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Watson, K.A, Tsitsanou, K.E, Gregoriou, M, Zographos, S.E, Skamnaki, V.T, Oikonomakos, N.G, Fleet, G.W, Johnson, L.N. | | Deposit date: | 2000-09-08 | | Release date: | 2000-10-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Kinetic and crystallographic studies of glucopyranose spirohydantoin and glucopyranosylamine analogs inhibitors of glycogen phosphorylase.
Proteins, 61, 2005
|
|
