4QUC
 
 | | Crystal structure of chromodomain of Rhino | | Descriptor: | RE36324p | | Authors: | Li, S, Patel, D.J. | | Deposit date: | 2014-07-10 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Transgenerationally inherited piRNAs trigger piRNA biogenesis by changing the chromatin of piRNA clusters and inducing precursor processing.
Genes Dev., 28, 2014
|
|
4QUF
 
 | | crystal structure of chromodomain of Rhino with H3K9me3 | | Descriptor: | H3(1-15)K9me3 peptide, RE36324p | | Authors: | Li, S, Patel, D.J. | | Deposit date: | 2014-07-10 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Transgenerationally inherited piRNAs trigger piRNA biogenesis by changing the chromatin of piRNA clusters and inducing precursor processing.
Genes Dev., 28, 2014
|
|
3HKS
 
 | | Crystal structure of eukaryotic translation initiation factor eIF-5A2 from Arabidopsis thaliana | | Descriptor: | 1,2-ETHANEDIOL, Eukaryotic translation initiation factor 5A-2 | | Authors: | Teng, Y.B, He, Y.X, Jiang, Y.L, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2009-05-25 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Arabidopsis translation initiation factor eIF-5A2
Proteins, 77, 2009
|
|
4TLL
 
 | | Crystal structure of GluN1/GluN2B NMDA receptor, structure 1 | | Descriptor: | 1-AMINOCYCLOPROPANECARBOXYLIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[(1R,2S)-3-(4-benzylpiperidin-1-yl)-1-hydroxy-2-methylpropyl]phenol, ... | | Authors: | Gouaux, E, Lee, C.-H, Lu, W. | | Deposit date: | 2014-05-30 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | NMDA receptor structures reveal subunit arrangement and pore architecture.
Nature, 511, 2014
|
|
4TLM
 
 | | Crystal structure of GluN1/GluN2B NMDA receptor, structure 2 | | Descriptor: | 1-AMINOCYCLOPROPANECARBOXYLIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[(1R,2S)-3-(4-benzylpiperidin-1-yl)-1-hydroxy-2-methylpropyl]phenol, ... | | Authors: | Gouaux, E, Lee, C.-H, Lu, W. | | Deposit date: | 2014-05-30 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.77 Å) | | Cite: | NMDA receptor structures reveal subunit arrangement and pore architecture.
Nature, 511, 2014
|
|
7E5Y
 
 | | Molecular basis for neutralizing antibody 2B11 targeting SARS-CoV-2 RBD | | Descriptor: | 2B11 Fab Heavy chain, 2B11 Fab Light chain, Spike protein S1 | | Authors: | Wu, H, Yu, F, Wang, Q.S, Zhou, H, Wang, W.W, Zhao, T, Pan, Y.B, Yang, X.M. | | Deposit date: | 2021-02-21 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Screening of potent neutralizing antibodies against SARS-CoV-2 using convalescent patients-derived phage-display libraries.
Cell Discov, 7, 2021
|
|
4E78
 
 | |
2RQK
 
 | | NMR Solution Structure of Mesoderm Development (MESD) - closed conformation | | Descriptor: | Mesoderm development candidate 2 | | Authors: | Koehler, C, Lighthouse, J.K, Werther, T, Andersen, O.M, Diehl, A, Schmieder, P, Holdener, B.C, Oschkinat, H. | | Deposit date: | 2009-08-06 | | Release date: | 2009-08-25 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | The Structure of MESD45-184 Brings Light into the Mechanism of LDLR Family Folding
Structure, 19, 2011
|
|
2RQM
 
 | | NMR Solution Structure of Mesoderm Development (MESD) - open conformation | | Descriptor: | Mesoderm development candidate 2 | | Authors: | Koehler, C, Lighthouse, J.K, Werther, T, Andersen, O.M, Diehl, A, Schmieder, P, Holdener, B.C, Oschkinat, H. | | Deposit date: | 2009-08-14 | | Release date: | 2009-08-25 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | The Structure of MESD45-184 Brings Light into the Mechanism of LDLR Family Folding
Structure, 19, 2011
|
|
8F8V
 
 | | Crystal structure of Nb.X0 | | Descriptor: | Nb.X0 | | Authors: | Goldgur, Y, Ravetch, J, Gupta, A, Kao, K, Andi, B. | | Deposit date: | 2022-11-22 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Mechanism of glycoform specificity and in vivo protection by an anti-afucosylated IgG nanobody.
Nat Commun, 14, 2023
|
|
8F8W
 
 | | Crystal structure of Nb.X0 bound to the afucosylated human IgG1 fragment crystal form I | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nb.X0, afucosylated IgG1 fragment | | Authors: | Goldgur, Y, Ravetch, J, Gupta, A, Kao, K, Oren, D. | | Deposit date: | 2022-11-22 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Mechanism of glycoform specificity and in vivo protection by an anti-afucosylated IgG nanobody.
Nat Commun, 14, 2023
|
|
8F8X
 
 | | Crystal structure of Nb.X0 bound to the afucosylated human IgG1 fragment crystal form II | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nb.X0, Uncharacterized protein DKFZp686C11235 | | Authors: | Goldgur, Y, Ravetch, J, Gupta, A, Kao, K, Oren, D. | | Deposit date: | 2022-11-22 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mechanism of glycoform specificity and in vivo protection by an anti-afucosylated IgG nanobody.
Nat Commun, 14, 2023
|
|
7Y9G
 
 | | Crystal structure of diterpene synthase VenA from Streptomyces venezuelae ATCC 15439 in complex with pyrophosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhang, L.L, Xie, Z.Z, Huang, J.-W, Hu, Y.C, Liu, W.D, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-06-24 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Molecular insights into the catalytic promiscuity of a bacterial diterpene synthase.
Nat Commun, 14, 2023
|
|
7Y9H
 
 | | Crystal structure of diterpene synthase VenA from Streptomyces venezuelae ATCC 15439 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Diterpene synthase VenA, ... | | Authors: | Zhang, L.L, Xie, Z.Z, Huang, J.-W, Hu, Y.C, Liu, W.D, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-06-24 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Molecular insights into the catalytic promiscuity of a bacterial diterpene synthase.
Nat Commun, 14, 2023
|
|
3ZG6
 
 | | The novel de-long chain fatty acid function of human sirt6 | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, GLYCEROL, NAD-DEPENDENT PROTEIN DEACETYLASE SIRTUIN-6, ... | | Authors: | Wang, Y, Hao, Q. | | Deposit date: | 2012-12-15 | | Release date: | 2013-04-03 | | Last modified: | 2013-04-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Sirt6 Regulates Tnf-Alpha Secretion Through Hydrolysis of Long-Chain Fatty Acyl Lysine
Nature, 496, 2013
|
|
4KJT
 
 | | Structure of the L100F MUTANT OF DEHALOPEROXIDASE-HEMOGLOBIN A FROM AMPHITRITE ORNATA WITH OXYGEN | | Descriptor: | 1,2-ETHANEDIOL, Dehaloperoxidase A, OXYGEN MOLECULE, ... | | Authors: | Wang, C, Lovelace, L, Lebioda, L. | | Deposit date: | 2013-05-03 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Influence of heme environment structure on dioxygen affinity for the dual function Amphitrite ornata hemoglobin/dehaloperoxidase. Insights into the evolutional structure-function adaptations.
Arch.Biochem.Biophys., 545, 2014
|
|
5U36
 
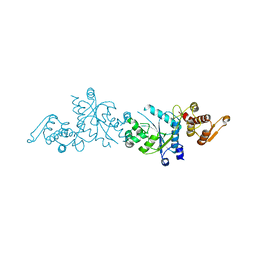 | | Crystal Structure Of A Mutant M. Jannashii Tyrosyl-tRNA Synthetase | | Descriptor: | Tyrosine--tRNA ligase | | Authors: | Luo, X, Fu, G, Zhu, X, Wilson, I.A, Wang, F. | | Deposit date: | 2016-12-01 | | Release date: | 2017-06-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Genetically encoding phosphotyrosine and its nonhydrolyzable analog in bacteria.
Nat. Chem. Biol., 13, 2017
|
|
8DGG
 
 | | Structure of glycosylated LAG-3 homodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Silberstein, J.L, Mathews, I.I, Frank, J.A, Chan, K.-W, Fernandez, D, Du, J, Wang, J, Kong, X.-P, Cochran, J.R. | | Deposit date: | 2022-06-23 | | Release date: | 2022-08-17 | | Last modified: | 2022-08-31 | | Method: | X-RAY DIFFRACTION (3.78 Å) | | Cite: | Structural basis for LAG-3 dimeric association and inhibition of T cell function
To Be Published
|
|
7E9G
 
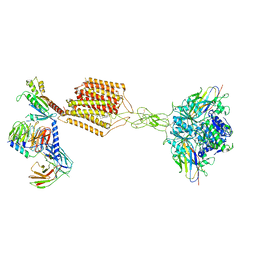 | | Cryo-EM structure of Gi-bound metabotropic glutamate receptor mGlu2 | | Descriptor: | (1S,2S,5R,6S)-2-aminobicyclo[3.1.0]hexane-2,6-dicarboxylic acid, 1-butyl-3-chloranyl-4-(4-phenylpiperidin-1-yl)pyridin-2-one, DN13, ... | | Authors: | Lin, S, Han, S, Zhao, Q, Wu, B. | | Deposit date: | 2021-03-04 | | Release date: | 2021-06-23 | | Last modified: | 2021-08-04 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of G i -bound metabotropic glutamate receptors mGlu2 and mGlu4.
Nature, 594, 2021
|
|
7E9H
 
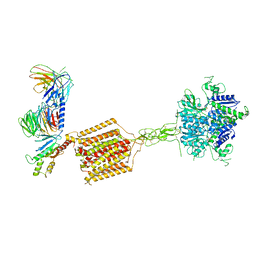 | | Cryo-EM structure of Gi-bound metabotropic glutamate receptor mGlu4 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-3, ... | | Authors: | Lin, S, Han, S, Zhao, Q, Wu, B. | | Deposit date: | 2021-03-04 | | Release date: | 2021-06-23 | | Last modified: | 2021-07-07 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of G i -bound metabotropic glutamate receptors mGlu2 and mGlu4.
Nature, 594, 2021
|
|
4F4U
 
 | |
4F56
 
 | | The bicyclic intermediate structure provides insights into the desuccinylation mechanism of SIRT5 | | Descriptor: | 3-[(2R,3aR,5R,6R,6aR)-5-({[(S)-{[(S)-{[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}methyl)-2,6-dihydroxytetrahydrofuro[2,3-d][1,3]oxathiol-2-yl]propanoic acid, NAD-dependent lysine demalonylase and desuccinylase sirtuin-5, mitochondrial, ... | | Authors: | Zhou, Y, Hao, Q. | | Deposit date: | 2012-05-11 | | Release date: | 2012-06-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Bicyclic Intermediate Structure Provides Insights into the Desuccinylation Mechanism of Human Sirtuin 5 (SIRT5)
J.Biol.Chem., 287, 2012
|
|
6MOA
 
 | | C-terminal bromodomain of human BRD2 in complex with 4-(2-cyclopropyl-7-(6-methylquinolin-5-yl)-1H-benzo[d]imidazol-5-yl)-3,5-dimethylisoxazole inhibitor | | Descriptor: | 4-(2-cyclopropyl-7-(6-methylquinolin-5-yl)-1H-benzo[d]imidazol-5-yl)-3,5-dimethylisoxazole, Bromodomain-containing protein 2, GLYCEROL | | Authors: | Lansdon, E.B, Newby, Z.E.R. | | Deposit date: | 2018-10-04 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.271 Å) | | Cite: | Structure-guided discovery of a novel, potent, and orally bioavailable 3,5-dimethylisoxazole aryl-benzimidazole BET bromodomain inhibitor.
Bioorg. Med. Chem., 27, 2019
|
|
6MO7
 
 | | N-terminal bromodomain of human BRD2 with N-((4-(3-(N-cyclopentylsulfamoyl)-4-methylphenyl)-3-methylisoxazol-5-yl)methyl)acetamide inhibitor | | Descriptor: | Bromodomain-containing protein 2, N-({4-[3-(cyclopentylsulfamoyl)-4-methylphenyl]-3-methyl-1,2-oxazol-5-yl}methyl)acetamide | | Authors: | Lansdon, E.B, Newby, Z.E.R. | | Deposit date: | 2018-10-04 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-guided discovery of a novel, potent, and orally bioavailable 3,5-dimethylisoxazole aryl-benzimidazole BET bromodomain inhibitor.
Bioorg. Med. Chem., 27, 2019
|
|
6MO9
 
 | | N-terminal bromodomain of human BRD2 in complex with N-cyclopentyl-7-(3,5-dimethylisoxazol-4-yl)quinoline-5-sulfonamide inhibitor | | Descriptor: | Bromodomain-containing protein 2, N-cyclopentyl-7-(3,5-dimethyl-1,2-oxazol-4-yl)quinoline-5-sulfonamide | | Authors: | Lansdon, E.B, Newby, Z.E.R. | | Deposit date: | 2018-10-04 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structure-guided discovery of a novel, potent, and orally bioavailable 3,5-dimethylisoxazole aryl-benzimidazole BET bromodomain inhibitor.
Bioorg. Med. Chem., 27, 2019
|
|
