4DNU
 
 | | Crystal structure of the W285A mutant of UVB-resistance protein UVR8 | | 分子名称: | AT5g63860/MGI19_6 | | 著者 | Wu, D, Hu, Q, Yan, Z, Chen, W, Yan, C, Zhang, J, Wang, J, Shi, Y. | | 登録日 | 2012-02-09 | | 公開日 | 2012-03-07 | | 最終更新日 | 2013-07-17 | | 実験手法 | X-RAY DIFFRACTION (1.764 Å) | | 主引用文献 | Structural basis of ultraviolet-B perception by UVR8.
Nature, 484, 2012
|
|
4I51
 
 | | Methyltransferase domain of HUMAN EUCHROMATIC HISTONE METHYLTRANSFERASE 1, mutant Y1211A | | 分子名称: | GLYCEROL, H3K9 NE-ALLYL PEPTIDE, Histone-lysine N-methyltransferase EHMT1, ... | | 著者 | Dong, A, Zeng, H, Walker, J.R, Islam, K, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Lou, M, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | 登録日 | 2012-11-28 | | 公開日 | 2012-12-19 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Defining efficient enzyme-cofactor pairs for bioorthogonal profiling of protein methylation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
8JJ9
 
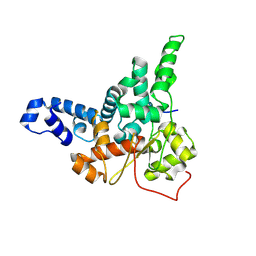 | |
5HGU
 
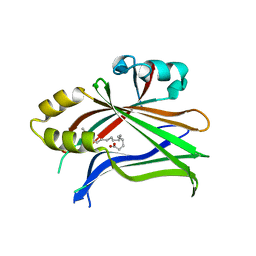 | |
4C13
 
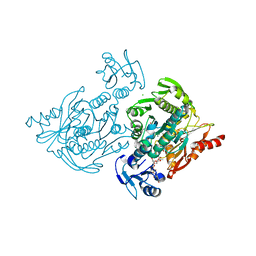 | | x-ray crystal structure of Staphylococcus aureus MurE with UDP-MurNAc- Ala-Glu-Lys | | 分子名称: | CHLORIDE ION, MAGNESIUM ION, PHOSPHATE ION, ... | | 著者 | Ruane, K.M, Roper, D.I, Fulop, V, Barreteau, H, Boniface, A, Dementin, S, Blanot, D, Mengin-Lecreulx, D, Gobec, S, Dessen, A, Dowson, C.G, Lloyd, A.J. | | 登録日 | 2013-08-09 | | 公開日 | 2013-10-02 | | 最終更新日 | 2021-03-17 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Discovery of a first-in-class CDK2 selective degrader for AML differentiation therapy.
Nat.Chem.Biol., 2021
|
|
5EIB
 
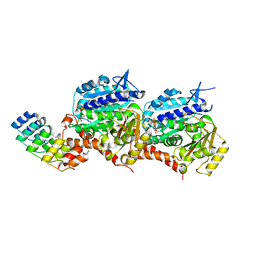 | |
2ISC
 
 | | Crystal structure of Purine Nucleoside Phosphorylase from Trichomonas vaginalis with DADMe-Imm-A | | 分子名称: | (3R,4R)-1-[(4-AMINO-5H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)METHYL]-4-(HYDROXYMETHYL)PYRROLIDIN-3-OL, PHOSPHATE ION, purine nucleoside phosphorylase | | 著者 | Rinaldo-Matthis, A, Almo, S.C, Schramm, V.L. | | 登録日 | 2006-10-17 | | 公開日 | 2007-06-05 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Inhibition and structure of Trichomonas vaginalis purine nucleoside phosphorylase with picomolar transition state analogues
Biochemistry, 46, 2007
|
|
6L8O
 
 | |
6L8N
 
 | | Crystal structure of the K. lactis Rad5 | | 分子名称: | DNA repair protein RAD5, ZINC ION | | 著者 | Shen, M, Xiang, S. | | 登録日 | 2019-11-06 | | 公開日 | 2020-11-11 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Structural basis for the multi-activity factor Rad5 in replication stress tolerance.
Nat Commun, 12, 2021
|
|
5CI6
 
 | |
2LWF
 
 | | Structure of N-terminal domain of a plant Grx | | 分子名称: | Monothiol glutaredoxin-S16, chloroplastic | | 著者 | Feng, Y. | | 登録日 | 2012-07-28 | | 公開日 | 2013-05-22 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural insights into the N-terminal GIY-YIG endonuclease activity of Arabidopsis glutaredoxin AtGRXS16 in chloroplasts.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4NCM
 
 | |
4NCE
 
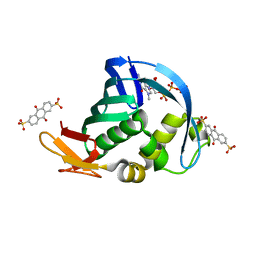 | | Influenza polymerase basic protein 2 (PB2) bound to 7-methyl-GTP | | 分子名称: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, 9,10-dioxo-9,10-dihydroanthracene-2,6-disulfonic acid, Polymerase basic protein 2 | | 著者 | Jacobs, M.D. | | 登録日 | 2013-10-24 | | 公開日 | 2014-07-30 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Discovery of a Novel, First-in-Class, Orally Bioavailable Azaindole Inhibitor (VX-787) of Influenza PB2.
J.Med.Chem., 57, 2014
|
|
2I4T
 
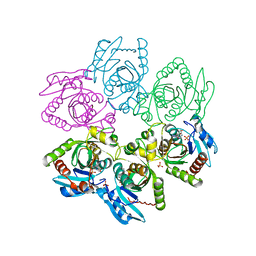 | | Crystal structure of Purine Nucleoside Phosphorylase from Trichomonas vaginalis with Imm-A | | 分子名称: | 3,4-PYRROLIDINEDIOL,2-(4-AMINO-5H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)-2S,3S,4R,5R, PHOSPHATE ION, Trichomonas vaginalis purine nucleoside phosphorylase | | 著者 | Rinaldo-Matthis, A, Schramm, V.L, Almo, S.C. | | 登録日 | 2006-08-22 | | 公開日 | 2007-06-05 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.74 Å) | | 主引用文献 | Inhibition and structure of Trichomonas vaginalis purine nucleoside phosphorylase with picomolar transition state analogues.
Biochemistry, 46, 2007
|
|
4P1U
 
 | |
3U96
 
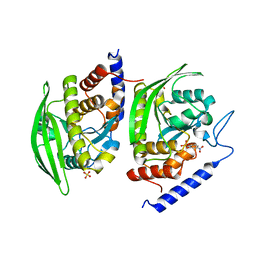 | | Crystal Structure of YopHQ357F(Catalytic Domain, Residues 163-468) in complex with pNCS | | 分子名称: | N,4-DIHYDROXY-N-OXO-3-(SULFOOXY)BENZENAMINIUM, SULFATE ION, Tyrosine-protein phosphatase yopH | | 著者 | Ho, M.C, Ke, S. | | 登録日 | 2011-10-17 | | 公開日 | 2012-08-29 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Investigation of catalytic loop structure, dynamics, and function relationship of Yersinia protein tyrosine phosphatase by temperature-jump relaxation spectroscopy and X-ray structural determination.
J.Phys.Chem.B, 116, 2012
|
|
6K15
 
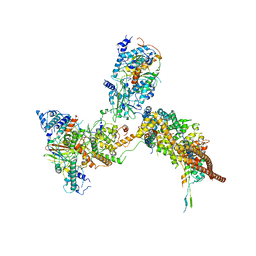 | | RSC substrate-recruitment module | | 分子名称: | Chromatin structure-remodeling complex protein RSC3, Chromatin structure-remodeling complex protein RSC30, Chromatin structure-remodeling complex protein RSC58, ... | | 著者 | Ye, Y.P, Wu, H, Chen, K.J, Verma, N, Cairns, B, Gao, N, Chen, Z.C. | | 登録日 | 2019-05-09 | | 公開日 | 2019-11-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structure of the RSC complex bound to the nucleosome.
Science, 366, 2019
|
|
6KW3
 
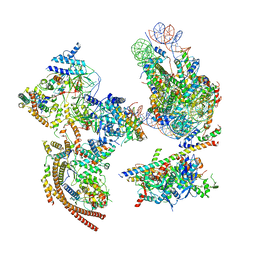 | | The ClassA RSC-Nucleosome Complex | | 分子名称: | Actin-like protein ARP9, Actin-related protein 7, Chromatin structure-remodeling complex protein RSC3, ... | | 著者 | Ye, Y.P, Wu, H, Chen, K.J, Verma, N, Cairns, B, Gao, N, Chen, Z.C. | | 登録日 | 2019-09-05 | | 公開日 | 2019-11-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (7.13 Å) | | 主引用文献 | Structure of the RSC complex bound to the nucleosome.
Science, 366, 2019
|
|
6KYF
 
 | | Crystal structure of an anti-CRISPR protein | | 分子名称: | AcrF11, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Niu, Y, Wang, H, Zhang, Y, Feng, Y. | | 登録日 | 2019-09-18 | | 公開日 | 2020-09-23 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (3.07 Å) | | 主引用文献 | A Type I-F Anti-CRISPR Protein Inhibits the CRISPR-Cas Surveillance Complex by ADP-Ribosylation.
Mol.Cell, 80, 2020
|
|
6KW4
 
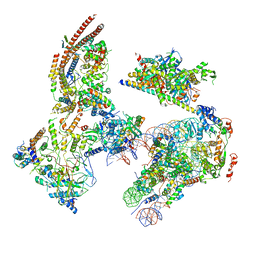 | | The ClassB RSC-Nucleosome Complex | | 分子名称: | Actin-like protein ARP9, Actin-related protein 7, Chromatin structure-remodeling complex protein RSC3, ... | | 著者 | Ye, Y.P, Wu, H, Chen, K.J, Verma, N, Cairns, B, Gao, N, Chen, Z.C. | | 登録日 | 2019-09-06 | | 公開日 | 2019-11-13 | | 最終更新日 | 2023-07-26 | | 実験手法 | ELECTRON MICROSCOPY (7.55 Å) | | 主引用文献 | Structure of the RSC complex bound to the nucleosome.
Science, 366, 2019
|
|
8ITE
 
 | |
3J3W
 
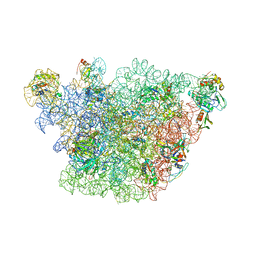 | | Atomic model of the immature 50S subunit from Bacillus subtilis (state II-a) | | 分子名称: | 50S ribosomal protein L1, 50S ribosomal protein L11, 50S ribosomal protein L13, ... | | 著者 | Li, N, Guo, Q, Zhang, Y, Yuan, Y, Ma, C, Lei, J, Gao, N. | | 登録日 | 2013-04-28 | | 公開日 | 2013-06-12 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (10.7 Å) | | 主引用文献 | Cryo-EM structures of the late-stage assembly intermediates of the bacterial 50S ribosomal subunit
Nucleic Acids Res., 41, 2013
|
|
3K6Y
 
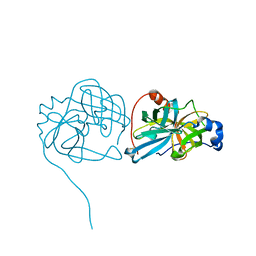 | | Crystal structure of Rv3671c protease from M. tuberculosis, active form | | 分子名称: | POSSIBLE MEMBRANE-ASSOCIATED SERINE PROTEASE | | 著者 | Biswas, T, Small, J, Vandal, O, Ehrt, S, Tsodikov, O.V. | | 登録日 | 2009-10-10 | | 公開日 | 2010-10-13 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Structural insight into serine protease Rv3671c that Protects M. tuberculosis from oxidative and acidic stress.
Structure, 18, 2010
|
|
3J3V
 
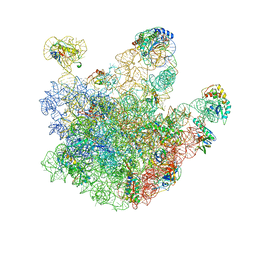 | | Atomic model of the immature 50S subunit from Bacillus subtilis (state I-a) | | 分子名称: | 50S ribosomal protein L1, 50S ribosomal protein L11, 50S ribosomal protein L13, ... | | 著者 | Li, N, Guo, Q, Zhang, Y, Yuan, Y, Ma, C, Lei, J, Gao, N. | | 登録日 | 2013-04-28 | | 公開日 | 2013-06-12 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (13.3 Å) | | 主引用文献 | Cryo-EM structures of the late-stage assembly intermediates of the bacterial 50S ribosomal subunit
Nucleic Acids Res., 41, 2013
|
|
3LT3
 
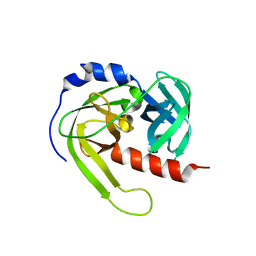 | | Crystal structure of Rv3671c from M. tuberculosis H37Rv, Ser343Ala mutant, inactive form | | 分子名称: | POSSIBLE MEMBRANE-ASSOCIATED SERINE PROTEASE | | 著者 | Biswas, T, Small, J, Vandal, O, Ehrt, S, Tsodikov, O.V. | | 登録日 | 2010-02-14 | | 公開日 | 2010-11-03 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural insight into serine protease Rv3671c that Protects M. tuberculosis from oxidative and acidic stress.
Structure, 18, 2010
|
|
