5JH5
 
 | | Structural Basis for the Hierarchical Assembly of the Core of PRC1.1 | | Descriptor: | BCL-6 corepressor-like protein 1, Lysine-specific demethylase 2B, Polycomb group RING finger protein 1, ... | | Authors: | Wong, S.J, Taylor, A.B, Hart, P.J, Kim, C.A. | | Deposit date: | 2016-04-20 | | Release date: | 2016-09-14 | | Last modified: | 2016-10-19 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | KDM2B Recruitment of the Polycomb Group Complex, PRC1.1, Requires Cooperation between PCGF1 and BCORL1.
Structure, 24, 2016
|
|
5JIE
 
 | |
5L1M
 
 | | CASKIN2 SAM domain tandem | | Descriptor: | Caskin-2 | | Authors: | Donaldson, L.W, Kwan, J.J, Saridakis, V. | | Deposit date: | 2016-07-29 | | Release date: | 2016-08-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.751 Å) | | Cite: | A new mode of SAM domain mediated oligomerization observed in the CASKIN2 neuronal scaffolding protein.
Cell Commun. Signal, 14, 2016
|
|
6DR5
 
 | |
6DR4
 
 | |
6DR6
 
 | |
5CZ1
 
 | |
5D7U
 
 | | Crystal structure of the C-terminal domain of MMTV integrase | | Descriptor: | ISOPROPYL ALCOHOL, Pr160 | | Authors: | Cook, N.J, Pye, V.E, Ballandras-Colas, A, Engelman, A, Cherepanov, P. | | Deposit date: | 2015-08-14 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Cryo-EM reveals a novel octameric integrase structure for betaretroviral intasome function.
Nature, 530, 2016
|
|
5CZ2
 
 | | Crystal structure of a two-domain fragment of MMTV integrase | | Descriptor: | MAGNESIUM ION, Pol polyprotein, ZINC ION | | Authors: | Cook, N, Ballandras-Colas, A, Engelman, A, Cherepanov, P. | | Deposit date: | 2015-07-31 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Cryo-EM reveals a novel octameric integrase structure for betaretroviral intasome function.
Nature, 530, 2016
|
|
3C7N
 
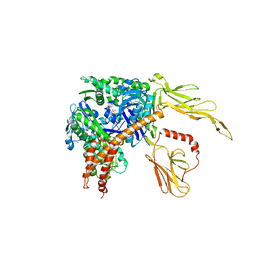 | | Structure of the Hsp110:Hsc70 Nucleotide Exchange Complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, CHLORIDE ION, ... | | Authors: | Schuermann, J.P, Jiang, J, Hart, P.J, Sousa, R. | | Deposit date: | 2008-02-07 | | Release date: | 2008-05-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.115 Å) | | Cite: | Structure of the Hsp110:Hsc70 nucleotide exchange machine
Mol.Cell, 31, 2008
|
|
4FQN
 
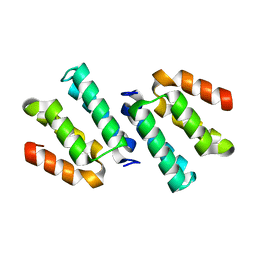 | | Crystal structure of the CCM2 C-terminal Harmonin Homology Domain (HHD) | | Descriptor: | Malcavernin | | Authors: | Fisher, O.S, Zhang, R, Li, X, Murphy, J.W, Boggon, T.J. | | Deposit date: | 2012-06-25 | | Release date: | 2012-12-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural studies of cerebral cavernous malformations 2 (CCM2) reveal a folded helical domain at its C-terminus.
Febs Lett., 587, 2013
|
|
4HPM
 
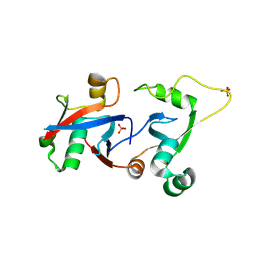 | | PCGF1 Ub fold (RAWUL)/BCORL1 PUFD Complex | | Descriptor: | BCL-6 corepressor-like protein 1, PHOSPHATE ION, Polycomb group RING finger protein 1 | | Authors: | Junco, S.E, Wang, R, Gaipa, J, Taylor, A.B, Gearhart, M.D, Bardwell, V.J, Hart, P.J, Kim, C.A. | | Deposit date: | 2012-10-24 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of the Polycomb Group Protein PCGF1 in Complex with BCOR Reveals Basis for Binding Selectivity of PCGF Homologs.
Structure, 21, 2013
|
|
4HPL
 
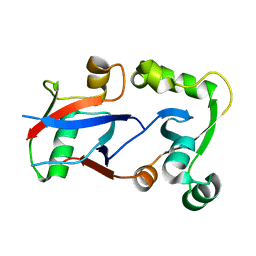 | | PCGF1 Ub fold (RAWUL)/BCOR PUFD Complex | | Descriptor: | BCL-6 corepressor, Polycomb group RING finger protein 1 | | Authors: | Junco, S.E, Wang, R, Gaipa, J, Taylor, A.B, Gearhart, M.D, Bardwell, V.J, Hart, P.J, Kim, C.A. | | Deposit date: | 2012-10-24 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Polycomb Group Protein PCGF1 in Complex with BCOR Reveals Basis for Binding Selectivity of PCGF Homologs.
Structure, 21, 2013
|
|
3K6B
 
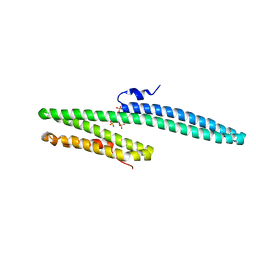 | | X-ray crystal structure of the E2 domain of APL-1 from C. elegans, in complex with sucrose octasulfate (SOS) | | Descriptor: | 2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, Beta-amyloid-like protein | | Authors: | Hoopes, J.T, Ha, Y. | | Deposit date: | 2009-10-08 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural characterization of the E2 domain of APL-1, a Caenorhabditis elegans homolog of human amyloid precursor protein, and its heparin binding site
J.Biol.Chem., 285, 2010
|
|
3K66
 
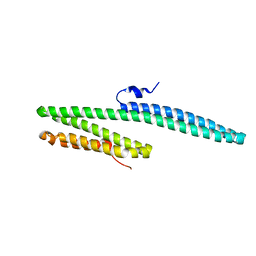 | | X-ray crystal structure of the E2 domain of C. elegans APL-1 | | Descriptor: | Beta-amyloid-like protein | | Authors: | Hoopes, J.T, Ha, Y. | | Deposit date: | 2009-10-08 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural characterization of the E2 domain of APL-1, a Caenorhabditis elegans homolog of human amyloid precursor protein, and its heparin binding site
J.Biol.Chem., 285, 2010
|
|
5VA4
 
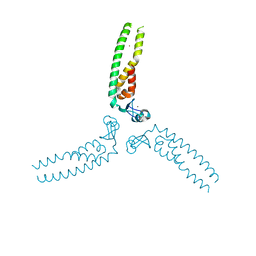 | |
3NYJ
 
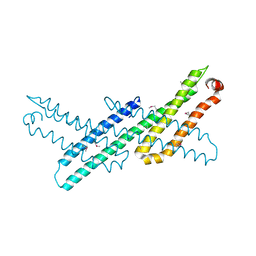 | | Crystal Structure Analysis of APP E2 domain | | Descriptor: | Amyloid beta A4 protein, OSMIUM ION | | Authors: | Ha, Y, Hu, J, Lee, S, Liu, X. | | Deposit date: | 2010-07-15 | | Release date: | 2011-06-01 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The E2 Domains of APP and APLP1 Share a Conserved Mode of Dimerization.
Biochemistry, 50, 2011
|
|
6BMC
 
 | | The structure of a dimeric type II DAH7PS associated with pyocyanin biosynthesis in Pseudomonas aeruginosa | | Descriptor: | CHLORIDE ION, COBALT (II) ION, PHOSPHOENOLPYRUVATE, ... | | Authors: | Sterritt, O.W, Jameson, G.B, Parker, E.J. | | Deposit date: | 2017-11-14 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and functional characterisation of the entry point to pyocyanin biosynthesis inPseudomonas aeruginosadefines a new 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase subclass.
Biosci. Rep., 38, 2018
|
|
2Q8T
 
 | | Crystal Structure of the CC chemokine CCL14 | | Descriptor: | CCL14 | | Authors: | Blain, K.Y. | | Deposit date: | 2007-06-11 | | Release date: | 2007-09-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural and Functional Characterization of CC Chemokine CCL14
Biochemistry, 46, 2007
|
|
2Q8R
 
 | |
2QSQ
 
 | | Crystal structure of the N-terminal domain of carcinoembryonic antigen (CEA) | | Descriptor: | CHLORIDE ION, Carcinoembryonic antigen-related cell adhesion molecule 5, GLYCEROL | | Authors: | Le Trong, I, Korotkova, N, Moseley, S.L, Stenkamp, R.E. | | Deposit date: | 2007-07-31 | | Release date: | 2008-01-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Binding of Dr adhesins of Escherichia coli to carcinoembryonic antigen triggers receptor dissociation.
Mol.Microbiol., 67, 2008
|
|
2QST
 
 | |
3SLK
 
 | |
1VF6
 
 | | 2.1 Angstrom crystal structure of the PALS-1-L27N and PATJ L27 heterodimer complex | | Descriptor: | MAGUK p55 subfamily member 5, PALS1-associated tight junction protein | | Authors: | Li, Y, Lavie, A, Margolis, B, Karnak, D. | | Deposit date: | 2004-04-09 | | Release date: | 2004-04-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for L27 domain-mediated assembly of signaling and cell polarity complexes.
Embo J., 23, 2004
|
|
4QNB
 
 | |
