7EZC
 
 | | Adenosine A2a receptor mutant-I92N | | Descriptor: | 6-(2,2-diphenylethylamino)-9-[(2R,3R,4S,5S)-5-(ethylcarbamoyl)-3,4-dihydroxy-oxolan-2-yl]-N-[2-[(1-pyridin-2-ylpiperidin-4-yl)carbamoylamino]ethyl]purine-2-carboxamide, Adenosine receptor A2a,Soluble cytochrome b562 | | Authors: | Cui, M, Zhou, Q, Yao, D, Zhao, S, Song, G. | | Deposit date: | 2021-06-01 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Crystal structure of a constitutive active mutant of adenosine A 2A receptor.
Iucrj, 9, 2022
|
|
7N6B
 
 | | Structure of MmpL3 reconstituted into lipid nanodisc in the TMM bound state | | Descriptor: | 6-O-[(2S)-2-{(1S)-18-[(1R,2R)-2-hexylcyclopropyl]-1-hydroxyoctadecyl}tricosanoyl]-alpha-D-glucopyranosyl alpha-D-glucopyranoside, MmpL3 transporter | | Authors: | Su, C.C, Yu, E. | | Deposit date: | 2021-06-08 | | Release date: | 2021-09-01 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structures of the mycobacterial membrane protein MmpL3 reveal its mechanism of lipid transport.
Plos Biol., 19, 2021
|
|
6KZQ
 
 | | structure of PTP-MEG2 and NSF-pY83 peptide complex | | Descriptor: | NSF-pY83 peptide, Tyrosine-protein phosphatase non-receptor type 9 | | Authors: | Xu, Y.F, Chen, X, Yu, X, Sun, J.P. | | Deposit date: | 2019-09-25 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | PTP-MEG2 regulates quantal size and fusion pore opening through two distinct structural bases and substrates.
Embo Rep., 22, 2021
|
|
6L03
 
 | | structure of PTP-MEG2 and MUNC18-1-pY145 peptide complex | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 9, stxbp1-pY145 peptide | | Authors: | Xu, Y.F, Chen, X, Yu, X, Sun, J.P. | | Deposit date: | 2019-09-25 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.084 Å) | | Cite: | PTP-MEG2 regulates quantal size and fusion pore opening through two distinct structural bases and substrates.
Embo Rep., 22, 2021
|
|
7MXO
 
 | | CryoEM structure of human NKCC1 | | Descriptor: | CHLORIDE ION, POTASSIUM ION, Solute carrier family 12 member 2 | | Authors: | Moseng, M.A. | | Deposit date: | 2021-05-19 | | Release date: | 2022-09-28 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Inhibition mechanism of NKCC1 involves the carboxyl terminus and long-range conformational coupling.
Sci Adv, 8, 2022
|
|
7N3N
 
 | | CryoEM structure of human NKCC1 state Fu-I | | Descriptor: | 5-(AMINOSULFONYL)-4-CHLORO-2-[(2-FURYLMETHYL)AMINO]BENZOIC ACID, Solute carrier family 12 member 2 | | Authors: | Moseng, M.A. | | Deposit date: | 2021-06-01 | | Release date: | 2022-09-28 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Inhibition mechanism of NKCC1 involves the carboxyl terminus and long-range conformational coupling.
Sci Adv, 8, 2022
|
|
7SFL
 
 | | Human NKCC1 state Fu-II | | Descriptor: | 5-(AMINOSULFONYL)-4-CHLORO-2-[(2-FURYLMETHYL)AMINO]BENZOIC ACID, Solute carrier family 12 member 2 | | Authors: | Moseng, M.A. | | Deposit date: | 2021-10-04 | | Release date: | 2022-09-28 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Inhibition mechanism of NKCC1 involves the carboxyl terminus and long-range conformational coupling.
Sci Adv, 8, 2022
|
|
7SMP
 
 | | CryoEM structure of NKCC1 Bu-I | | Descriptor: | 3-(butylamino)-4-phenoxy-5-sulfamoylbenzoic acid, POTASSIUM ION, Solute carrier family 12 member 2 | | Authors: | Moseng, M.A. | | Deposit date: | 2021-10-26 | | Release date: | 2022-09-28 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Inhibition mechanism of NKCC1 involves the carboxyl terminus and long-range conformational coupling.
Sci Adv, 8, 2022
|
|
8STE
 
 | | Cryo-EM structure of NKCC1 Fu_CTD | | Descriptor: | 5-(AMINOSULFONYL)-4-CHLORO-2-[(2-FURYLMETHYL)AMINO]BENZOIC ACID, Solute carrier family 12 member 2 | | Authors: | Moseng, M.A. | | Deposit date: | 2023-05-10 | | Release date: | 2023-05-17 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Inhibition mechanism of NKCC1 involves the carboxyl terminus and long-range conformational coupling.
Sci Adv, 8, 2022
|
|
6CZQ
 
 | |
4XZZ
 
 | | Structure of Helicobacter pylori Csd6 in the ligand-free state | | Descriptor: | Conserved hypothetical secreted protein, GLYCEROL | | Authors: | Kim, H.S, Im, H.N, Yoon, H.J, Suh, S.W. | | Deposit date: | 2015-02-05 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The Cell Shape-determining Csd6 Protein from Helicobacter pylori Constitutes a New Family of l,d-Carboxypeptidase
J.Biol.Chem., 290, 2015
|
|
4Y4V
 
 | | Structure of Helicobacter pylori Csd6 in the D-Ala-bound state | | Descriptor: | Conserved hypothetical secreted protein, D-ALANINE, GLYCEROL | | Authors: | Kim, H.S, Im, H.N, Yoon, H.J, Suh, S.W. | | Deposit date: | 2015-02-11 | | Release date: | 2015-09-02 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The Cell Shape-determining Csd6 Protein from Helicobacter pylori Constitutes a New Family of l,d-Carboxypeptidase
J.Biol.Chem., 290, 2015
|
|
5WBX
 
 | |
5WC5
 
 | |
8F4R
 
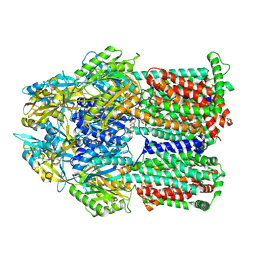 | | Gentamicin bound aminoglycoside efflux pump AcrD | | Descriptor: | (2R,3R,4R,5R)-2-((1S,2S,3R,4S,6R)-4,6-DIAMINO-3-((2R,3R,6S)-3-AMINO-6-(AMINOMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-2-HYDR OXYCYCLOHEXYLOXY)-5-METHYL-4-(METHYLAMINO)-TETRAHYDRO-2H-PYRAN-3,5-DIOL, Efflux pump membrane transporter | | Authors: | Zhang, Z. | | Deposit date: | 2022-11-11 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Cryo-EM Structures of AcrD Illuminate a Mechanism for Capturing Aminoglycosides from Its Central Cavity.
Mbio, 14, 2023
|
|
8F56
 
 | |
8F4N
 
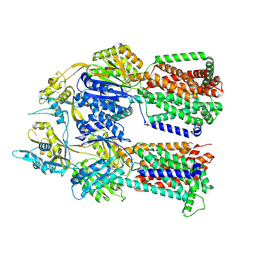 | | Dimer of aminoglycoside efflux pump AcrD | | Descriptor: | Efflux pump membrane transporter | | Authors: | Zhang, Z. | | Deposit date: | 2022-11-11 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Cryo-EM Structures of AcrD Illuminate a Mechanism for Capturing Aminoglycosides from Its Central Cavity.
Mbio, 14, 2023
|
|
8F3E
 
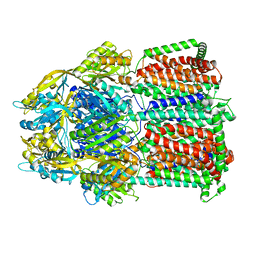 | | Trimer of aminoglycoside efflux pump AcrD | | Descriptor: | Efflux pump membrane transporter | | Authors: | Zhang, Z. | | Deposit date: | 2022-11-10 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Cryo-EM Structures of AcrD Illuminate a Mechanism for Capturing Aminoglycosides from Its Central Cavity.
Mbio, 14, 2023
|
|
4QNH
 
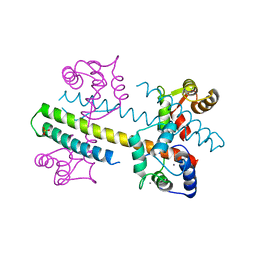 | | Calcium-calmodulin (T79D) complexed with the calmodulin binding domain from a small conductance potassium channel SK2-a | | Descriptor: | CALCIUM ION, Calmodulin, SULFATE ION, ... | | Authors: | Zhang, M, Pascal, J.M, Logothetis, D.E, Zhang, J.F. | | Deposit date: | 2014-06-17 | | Release date: | 2014-08-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Selective phosphorylation modulates the PIP2 sensitivity of the CaM-SK channel complex.
Nat.Chem.Biol., 10, 2014
|
|
7K8D
 
 | |
7K8A
 
 | |
7K8B
 
 | |
7K7M
 
 | | Crystal Structure of a membrane protein | | Descriptor: | Drug exporters of the RND superfamily-like protein, alpha-D-glucopyranose-(1-1)-6-O-decanoyl-alpha-D-glucopyranose | | Authors: | Su, C.-C. | | Deposit date: | 2020-09-23 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.33 Å) | | Cite: | Structures of the mycobacterial membrane protein MmpL3 reveal its mechanism of lipid transport.
Plos Biol., 19, 2021
|
|
7K8C
 
 | |
6JCS
 
 | | AAV5 in complex with AAVR | | Descriptor: | Capsid protein, Dyslexia-associated protein KIAA0319-like protein | | Authors: | Lou, Z, Zhang, R. | | Deposit date: | 2019-01-30 | | Release date: | 2019-08-14 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Divergent engagements between adeno-associated viruses with their cellular receptor AAVR.
Nat Commun, 10, 2019
|
|
