6QDL
 
 | |
6TV6
 
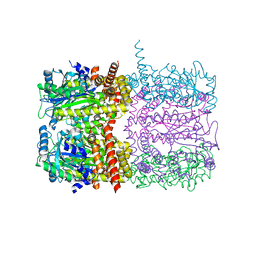 | | Octameric McsB from Bacillus subtilis. | | Descriptor: | MAGNESIUM ION, Protein-arginine kinase | | Authors: | Suskiewicz, M.J, Hajdusits, B, Meinhart, A, Clausen, T. | | Deposit date: | 2020-01-09 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | McsB forms a gated kinase chamber to mark aberrant bacterial proteins for degradation.
Elife, 10, 2021
|
|
8B9U
 
 | |
8B9O
 
 | |
8ATU
 
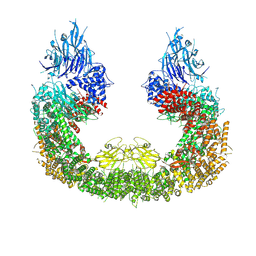 | | Cryo-EM structure of human BIRC6 | | Descriptor: | Baculoviral IAP repeat-containing protein 6, ZINC ION | | Authors: | Ehrmann, J.F, Grabarczyk, D.B, Clausen, T. | | Deposit date: | 2022-08-24 | | Release date: | 2023-02-15 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for regulation of apoptosis and autophagy by the BIRC6/SMAC complex.
Science, 379, 2023
|
|
8AUK
 
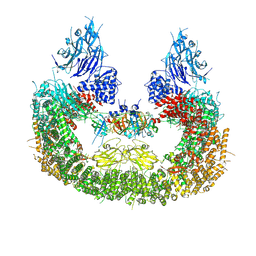 | | Cryo-EM structure of human BIRC6 in complex with HTRA2. | | Descriptor: | Baculoviral IAP repeat-containing protein 6, Serine protease HTRA2, mitochondrial, ... | | Authors: | Ehrmann, J.F, Grabarczyk, D.B, Clausen, T. | | Deposit date: | 2022-08-25 | | Release date: | 2023-02-15 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Structural basis for regulation of apoptosis and autophagy by the BIRC6/SMAC complex.
Science, 379, 2023
|
|
8AUW
 
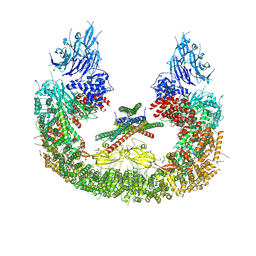 | |
8ATX
 
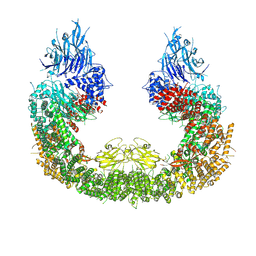 | |
1N31
 
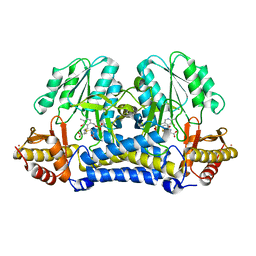 | | Structure of A Catalytically Inactive Mutant (K223A) of C-DES with a Substrate (Cystine) Linked to the Co-Factor | | Descriptor: | CYSTEINE, L-cysteine/cystine lyase C-DES, POTASSIUM ION, ... | | Authors: | Kaiser, J.T, Bruno, S, Clausen, T, Huber, R, Schiaretti, F, Mozzarelli, A, Kessler, D. | | Deposit date: | 2002-10-25 | | Release date: | 2003-01-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Snapshots of the Cystine Lyase "C-DES" during Catalysis: Studies in Solution and in the Crystalline State
J.Biol.Chem., 278, 2003
|
|
1N8P
 
 | | Crystal Structure of cystathionine gamma-lyase from yeast | | Descriptor: | Cystathionine gamma-lyase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Messerschmidt, A, Worbs, M, Steegborn, C, Wahl, M.C, Huber, R, Clausen, T. | | Deposit date: | 2002-11-21 | | Release date: | 2002-12-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Determinants of Enzymatic Specificity in the Cys-Met-Metabolism PLP-Dependent Enzymes Family: Crystal Structure of Cystathionine gamma-lyase from Yeast and Intrafamiliar Structural Comparison
BIOL.CHEM., 384, 2003
|
|
1POI
 
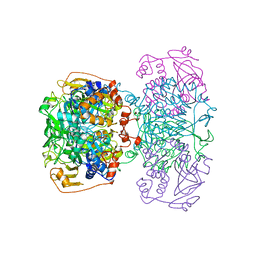 | | CRYSTAL STRUCTURE OF GLUTACONATE COENZYME A-TRANSFERASE FROM ACIDAMINOCOCCUS FERMENTANS TO 2.55 ANGSTOMS RESOLUTION | | Descriptor: | COPPER (II) ION, GLUTACONATE COENZYME A-TRANSFERASE | | Authors: | Jacob, U, Mack, M, Clausen, T, Huber, R, Buckel, W, Messerschmidt, A. | | Deposit date: | 1997-01-24 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Glutaconate CoA-transferase from Acidaminococcus fermentans: the crystal structure reveals homology with other CoA-transferases.
Structure, 5, 1997
|
|
1QGN
 
 | | CYSTATHIONINE GAMMA-SYNTHASE FROM NICOTIANA TABACUM | | Descriptor: | PROTEIN (CYSTATHIONINE GAMMA-SYNTHASE), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Steegborn, C, Messerschmidt, A, Laber, B, Streber, W, Huber, R, Clausen, T. | | Deposit date: | 1999-05-02 | | Release date: | 1999-08-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of cystathionine gamma-synthase from Nicotiana tabacum reveals its substrate and reaction specificity.
J.Mol.Biol., 290, 1999
|
|
1SOT
 
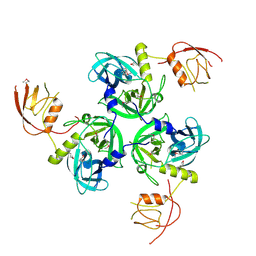 | | Crystal Structure of the DegS stress sensor | | Descriptor: | Protease degS | | Authors: | Wilken, C, Kitzing, K, Kurzbauer, R, Ehrmann, M, Clausen, T. | | Deposit date: | 2004-03-15 | | Release date: | 2004-06-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the DegS stress sensor: How a PDZ domain recognizes misfolded protein and activates a protease
Cell(Cambridge,Mass.), 117, 2004
|
|
7BII
 
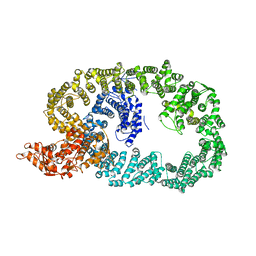 | | Crystal structure of Nematocida HUWE1 | | Descriptor: | E3 ubiquitin-protein ligase HUWE1 | | Authors: | Grabarczyk, D.B, Petrova, O.A, Meinhart, A, Kessler, D, Clausen, T. | | Deposit date: | 2021-01-12 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.037 Å) | | Cite: | HUWE1 employs a giant substrate-binding ring to feed and regulate its HECT E3 domain.
Nat.Chem.Biol., 17, 2021
|
|
3NWU
 
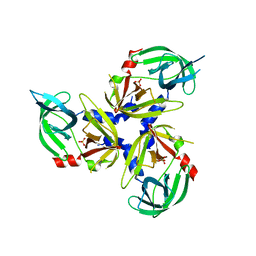 | | Substrate induced remodeling of the active site regulates HtrA1 activity | | Descriptor: | SULFATE ION, Serine protease HTRA1 | | Authors: | Truebestein, L, Tennstaedt, A, Hauske, P, Krojer, T, Kaiser, M, Clausen, T, Ehrmann, M. | | Deposit date: | 2010-07-11 | | Release date: | 2011-02-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Substrate-induced remodeling of the active site regulates human HTRA1 activity.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3NUM
 
 | | Substrate induced remodeling of the active site regulates HtrA1 activity | | Descriptor: | Serine protease HTRA1 | | Authors: | Truebestein, L, Tennstaedt, A, Hauske, P, Krojer, T, Kaiser, M, Clausen, T, Ehrmann, M. | | Deposit date: | 2010-07-07 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Substrate-induced remodeling of the active site regulates human HTRA1 activity.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3NZI
 
 | | Substrate induced remodeling of the active site regulates HtrA1 activity | | Descriptor: | Citrate synthase, Serine protease HTRA1 | | Authors: | Truebestein, L, Tennstaedt, A, Hauske, P, Krojer, T, Kaiser, M, Clausen, T, Ehrmann, M. | | Deposit date: | 2010-07-16 | | Release date: | 2011-02-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Substrate-induced remodeling of the active site regulates human HTRA1 activity.
Nat.Struct.Mol.Biol., 18, 2011
|
|
4A8D
 
 | | DegP dodecamer with bound OMP | | Descriptor: | OUTER MEMBRANE PROTEIN C, PERIPLASMIC SERINE ENDOPROTEASE DEGP | | Authors: | Malet, H, Krojer, T, Sawa, J, Schafer, E, Saibil, H.R, Ehrmann, M, Clausen, T. | | Deposit date: | 2011-11-20 | | Release date: | 2012-01-11 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (28 Å) | | Cite: | Newly Folded Substrates Inside the Molecular Cage of the Htra Chaperone Degq
Nat.Struct.Mol.Biol., 19, 2012
|
|
7AA4
 
 | | Structure of ClpC1-NTD bound to a CymA analogue | | Descriptor: | Negative regulator of genetic competence ClpC/mecB, polymer Cyclomarin A analogue | | Authors: | Meinhart, A, Morreale, F.E, Kaiser, M, Clausen, T. | | Deposit date: | 2020-09-03 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | BacPROTACs mediate targeted protein degradation in bacteria.
Cell, 185, 2022
|
|
7ABR
 
 | | Cryo-EM structure of B. subtilis ClpC (DWB mutant) hexamer bound to a substrate polypeptide | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Negative regulator of genetic competence ClpC/MecB, ... | | Authors: | Morreale, F.E, Meinhart, A, Haselbach, D, Clausen, T. | | Deposit date: | 2020-09-08 | | Release date: | 2021-10-06 | | Last modified: | 2022-07-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | BacPROTACs mediate targeted protein degradation in bacteria.
Cell, 185, 2022
|
|
6QDM
 
 | |
1SOZ
 
 | | Crystal Structure of DegS protease in complex with an activating peptide | | Descriptor: | Protease degS, activating peptide | | Authors: | Wilken, C, Kitzing, K, Kurzbauer, R, Ehrmann, M, Clausen, T. | | Deposit date: | 2004-03-16 | | Release date: | 2004-06-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the DegS stress sensor: How a PDZ domain recognizes misfolded protein and activates a protease
Cell(Cambridge,Mass.), 117, 2004
|
|
6QDK
 
 | |
3MH6
 
 | | HtrA proteases are activated by a conserved mechanism that can be triggered by distinct molecular cues | | Descriptor: | DIISOPROPYL PHOSPHONATE, Protease do | | Authors: | Krojer, T, Sawa, J, Huber, R, Clausen, T. | | Deposit date: | 2010-04-07 | | Release date: | 2010-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | HtrA proteases have a conserved activation mechanism that can be triggered by distinct molecular cues
Nat.Struct.Mol.Biol., 17, 2010
|
|
3MH4
 
 | |
