9JUE
 
 | |
9JQ8
 
 | |
8WT1
 
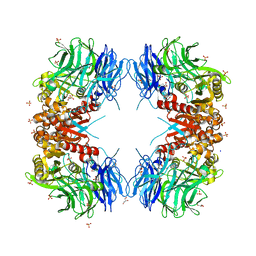 | | Crystal structure of S9 carboxypeptidase from Geobacillus sterothermophilus | | Descriptor: | ALANINE, CITRATE ANION, GLYCEROL, ... | | Authors: | Chandravanshi, K, Kumar, A, Sen, C, Singh, R, Bhange, G.B, Makde, R.D. | | Deposit date: | 2023-10-17 | | Release date: | 2024-03-13 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure and solution scattering of Geobacillus stearothermophilus S9 peptidase reveal structural adaptations for carboxypeptidase activity.
Febs Lett., 598, 2024
|
|
7YH4
 
 | |
8XZC
 
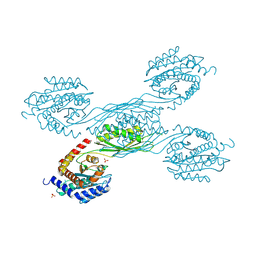 | |
1QC9
 
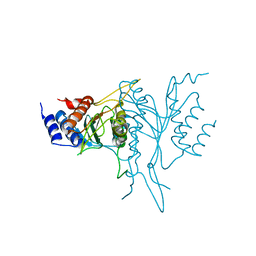 | |
4JDR
 
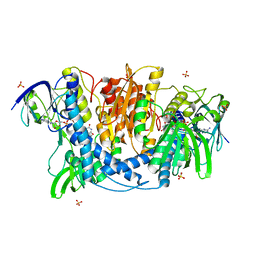 | | Dihydrolipoamide dehydrogenase of pyruvate dehydrogenase from escherichia coli | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Chandrasekhar, K, Arjunan, P, Furey, W. | | Deposit date: | 2013-02-25 | | Release date: | 2013-04-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insight to the Interaction of the Dihydrolipoamide Acetyltransferase (E2) Core with the Peripheral Components in the Escherichia coli Pyruvate Dehydrogenase Complex via Multifaceted Structural Approaches.
J.Biol.Chem., 288, 2013
|
|
2G67
 
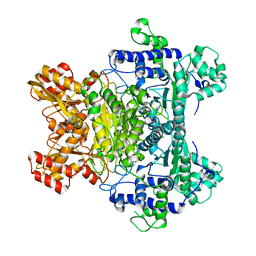 | |
4N72
 
 | |
6NHJ
 
 | | Atomic structures and deletion mutant reveal different capsid-binding patterns and functional significance of tegument protein pp150 in murine and human cytomegaloviruses with implications for therapeutic development | | Descriptor: | Major capsid protein, Minor capsid protein, Small capsomere-interacting protein, ... | | Authors: | Liu, W, Dai, X.H, Jih, J, Chan, K, Trang, P, Yu, X.K, Balogun, R, Mei, Y, Liu, F.Y, Zhou, Z.H. | | Deposit date: | 2018-12-22 | | Release date: | 2019-03-06 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Atomic structures and deletion mutant reveal different capsid-binding patterns and functional significance of tegument protein pp150 in murine and human cytomegaloviruses with implications for therapeutic development.
PLoS Pathog., 15, 2019
|
|
6WUQ
 
 | | Crystal structure of AjiA1 in apo form | | Descriptor: | AjiA1, MAGNESIUM ION, ZINC ION | | Authors: | Paiva, F.C.R, Chan, K, Leadlay, P, Dias, M.V.B. | | Deposit date: | 2020-05-05 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | The crystal structure of AjiA1 reveals a novel structural motion mechanism in the adenylate-forming enzyme family
Acta Crystallogr.,Sect.D, 76, 2020
|
|
6IRU
 
 | |
2MBL
 
 | | Solution NMR Structure of De novo designed Top7 Fold Protein Top7m13, Northeast Structural Genomics Consortium (NESG) Target OR33 | | Descriptor: | Top7 Fold Protein Top7m13 | | Authors: | Liu, G, Zanghellini, A.L, Chan, K, Xiao, R, Janjua, H, Kogan, S, Maglaqui, M, Ciccosanti, C, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-08-02 | | Release date: | 2013-11-13 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of De novo designed Top7 Fold Protein Top7m13, Northeast Structural Genomics Consortium (NESG) Target OR33
To be Published
|
|
2MBM
 
 | | Solution NMR Structure of De novo designed Top7 Fold Protein Top7m13, Northeast Structural Genomics Consortium (NESG) Target OR33 | | Descriptor: | Top7 Fold Protein Top7m13 | | Authors: | Liu, G, Zanghellini, A.L, Chan, K, Xiao, R, Janjua, H, Kogan, S, Maglaqui, M, Ciccosanti, C, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-08-02 | | Release date: | 2013-11-13 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of De novo designed Top7 Fold Protein Top7m13, Northeast Structural Genomics Consortium (NESG) Target OR33
To be Published
|
|
2N4E
 
 | | Solution NMR Structure of DE NOVO DESIGNED PROTEIN Top7NNSTYCC, Northeast Structural Genomics Consortium (NESG) Target OR34 | | Descriptor: | OR34 | | Authors: | Liu, G, Chan, K, Basanta, B, Xiao, R, Janjua, H, Kogan, S, Maglaqui, M, Ciccosanti, C, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-06-17 | | Release date: | 2015-12-09 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of DE NOVO DESIGNED PROTEIN Top7NNSTYCC, Northeast Structural Genomics Consortium (NESG) Target OR34
To be Published
|
|
2N41
 
 | | Solution NMR Structure of DE NOVO DESIGNED PROTEIN Top7NNSTYCC, Northeast Structural Genomics Consortium (NESG) Target OR34 | | Descriptor: | OR34 | | Authors: | Liu, G, Chan, K, Basanta, B, Xiao, R, Janjua, H, Kogan, S, Maglaqui, M, Ciccosanti, C, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-06-16 | | Release date: | 2016-03-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of DE NOVO DESIGNED PROTEIN Top7NNSTYCC, Northeast Structural Genomics Consortium (NESG) Target OR34
To be Published
|
|
2G25
 
 | | E. Coli Pyruvate Dehydrogenase Phosphonolactylthiamin Diphosphate Complex | | Descriptor: | 3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-2-{(1S)-1-HYDROXY-1-[(R)-HYDROXY(METHOXY)PHOSPHORYL]ETHYL}-5-(2-{[(S)-HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}ETHYL)-4-METHYL-1,3-THIAZOL-3-IUM, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Furey, W, Arjunan, P, Chandrasekhar, K. | | Deposit date: | 2006-02-15 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Thiamin-bound, Pre-decarboxylation Reaction Intermediate Analogue in the Pyruvate Dehydrogenase E1 Subunit Induces Large Scale Disorder-to-Order Transformations in the Enzyme and Reveals Novel Structural Features in the Covalently Bound Adduct.
J.Biol.Chem., 281, 2006
|
|
2G28
 
 | | E. Coli Pyruvate Dehydrogenase H407A variant Phosphonolactylthiamin Diphosphate Complex | | Descriptor: | 3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-2-{(1S)-1-HYDROXY-1-[(R)-HYDROXY(METHOXY)PHOSPHORYL]ETHYL}-5-(2-{[(S)-HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}ETHYL)-4-METHYL-1,3-THIAZOL-3-IUM, MAGNESIUM ION, Pyruvate dehydrogenase E1 component | | Authors: | Furey, W, Arjunan, P, Chandrasekhar, K. | | Deposit date: | 2006-02-15 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Thiamin-bound, Pre-decarboxylation Reaction Intermediate Analogue in the Pyruvate Dehydrogenase E1 Subunit Induces Large Scale Disorder-to-Order Transformations in the Enzyme and Reveals Novel Structural Features in the Covalently Bound Adduct.
J.Biol.Chem., 281, 2006
|
|
5JQD
 
 | | Antibody Fab Fragment | | Descriptor: | D80 Fab Fragment Heavy Chain, D80 Fab Fragment Light Chain | | Authors: | Zhang, Z, Prachanronarong, K, Gellatly, K, Marasco, W.A, Schiffer, C.A. | | Deposit date: | 2016-05-04 | | Release date: | 2017-11-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.591 Å) | | Cite: | Structural Basis of an Influenza Hemagglutinin Stem-Directed Antibody Retaining the G6 Idiotype
To Be Published
|
|
1GUJ
 
 | | Insulin at pH 2: structural analysis of the conditions promoting insulin fibre formation. | | Descriptor: | INSULIN, SULFATE ION | | Authors: | Whittingham, J.L, Scott, D.J, Chance, K, Wilson, A, Finch, J, Brange, J, Dodson, G.G. | | Deposit date: | 2002-01-28 | | Release date: | 2002-03-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Insulin at Ph2: Structural Analysis of the Conditions Promoting Insulin Fibre Formation
J.Mol.Biol., 318, 2002
|
|
5VK2
 
 | | Structural basis for antibody-mediated neutralization of Lassa virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hastie, K.M, Zandonatti, M.A, Kleinfelter, L.M, Rowland, M.L, Rowland, M.M, Chandra, K, Branco, L.M, Robinson, J.E, Garry, R.F, Saphire, E.O. | | Deposit date: | 2017-04-20 | | Release date: | 2017-05-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.201 Å) | | Cite: | Structural basis for antibody-mediated neutralization of Lassa virus.
Science, 356, 2017
|
|
4G2K
 
 | | Crystal structure of the Marburg Virus GP2 ectodomain in its post-fusion conformation | | Descriptor: | CHLORIDE ION, GLYCEROL, General control protein GCN4, ... | | Authors: | Malashkevich, V.N, Koellhoffer, J.F, Harrison, J.S, Toro, R, Bhosle, R.C, Chandran, K, Lai, J.R, Almo, S.C. | | Deposit date: | 2012-07-12 | | Release date: | 2012-09-12 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Marburg Virus GP2 Core Domain in Its Postfusion Conformation.
Biochemistry, 51, 2012
|
|
1RP7
 
 | | E. COLI PYRUVATE DEHYDROGENASE INHIBITOR COMPLEX | | Descriptor: | 2-{3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-4-METHYL-2-OXO-2,3-DIHYDRO-1,3-THIAZOL-5-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, MAGNESIUM ION, Pyruvate dehydrogenase E1 component | | Authors: | Arjunan, P, Chandrasekhar, K, Furey, W. | | Deposit date: | 2003-12-03 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural Determinants of Enzyme Binding Affinity: The E1 Component of Pyruvate Dehydrogenase from Escherichia coli in Complex with the Inhibitor Thiamin Thiazolone Diphosphate.
Biochemistry, 43, 2004
|
|
2MWQ
 
 | | Solution structure of PsbQ from spinacia oleracea | | Descriptor: | Oxygen-evolving enhancer protein 3, chloroplastic | | Authors: | Rathner, P, Mueller, N, Wimmer, R, Chandra, K. | | Deposit date: | 2014-11-19 | | Release date: | 2015-07-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR and molecular dynamics reveal a persistent alpha helix within the dynamic region of PsbQ from photosystem II of higher plants.
Proteins, 83, 2015
|
|
4UX3
 
 | | cohesin Smc3-HD:Scc1-N complex from yeast | | Descriptor: | MAGNESIUM ION, MITOTIC CHROMOSOME DETERMINANT-RELATED PROTEIN, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gligoris, T.G, Nasmyth, K, Lowe, J. | | Deposit date: | 2014-08-18 | | Release date: | 2014-12-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Closing the Cohesin Ring: Structure and Function of its Smc3-Kleisin Interface.
Science, 346, 2014
|
|
