6T90
 
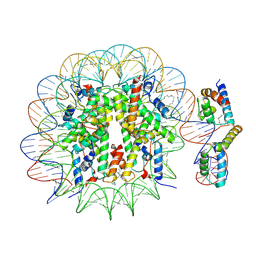 | | OCT4-SOX2-bound nucleosome - SHL-6 | | 分子名称: | DNA (146-MER), Green fluorescent protein,POU domain, class 5, ... | | 著者 | Michael, A.K, Kempf, G, Cavadini, S, Bunker, R.D, Thoma, N.H. | | 登録日 | 2019-10-25 | | 公開日 | 2020-05-06 | | 最終更新日 | 2020-07-08 | | 実験手法 | ELECTRON MICROSCOPY (3.05 Å) | | 主引用文献 | Mechanisms of OCT4-SOX2 motif readout on nucleosomes.
Science, 368, 2020
|
|
6T93
 
 | | Nucleosome with OCT4-SOX2 motif at SHL-6 | | 分子名称: | DNA (153-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | 著者 | Michael, A.K, Kempf, G, Cavadini, S, Bunker, R.D, Thoma, N.H. | | 登録日 | 2019-10-25 | | 公開日 | 2020-05-06 | | 最終更新日 | 2020-07-08 | | 実験手法 | ELECTRON MICROSCOPY (3.49 Å) | | 主引用文献 | Mechanisms of OCT4-SOX2 motif readout on nucleosomes.
Science, 368, 2020
|
|
8P83
 
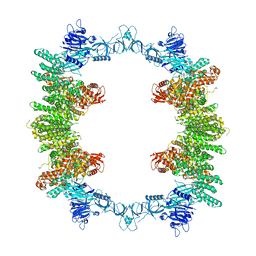 | | Cryo-EM structure of full-length human UBR5 (homotetramer) | | 分子名称: | E3 ubiquitin-protein ligase UBR5 | | 著者 | Aguirre, J.D, Kater, L, Kempf, G, Cavadini, S, Thoma, N.H. | | 登録日 | 2023-05-31 | | 公開日 | 2023-06-14 | | 最終更新日 | 2023-08-16 | | 実験手法 | ELECTRON MICROSCOPY (3.87 Å) | | 主引用文献 | UBR5 forms ligand-dependent complexes on chromatin to regulate nuclear hormone receptor stability.
Mol.Cell, 83, 2023
|
|
8OSL
 
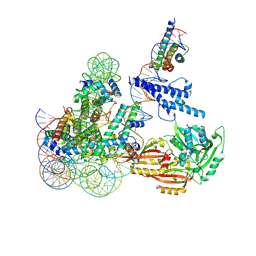 | | Cryo-EM structure of CLOCK-BMAL1 bound to the native Por enhancer nucleosome (map 2, additional 3D classification and flexible refinement) | | 分子名称: | Basic helix-loop-helix ARNT-like protein 1, Circadian locomoter output cycles protein kaput, DNA (147-MER), ... | | 著者 | Michael, A.K, Stoos, L, Kempf, G, Cavadini, S, Thoma, N. | | 登録日 | 2023-04-19 | | 公開日 | 2023-05-24 | | 最終更新日 | 2023-07-26 | | 実験手法 | ELECTRON MICROSCOPY (4.9 Å) | | 主引用文献 | Cooperation between bHLH transcription factors and histones for DNA access.
Nature, 619, 2023
|
|
8OTS
 
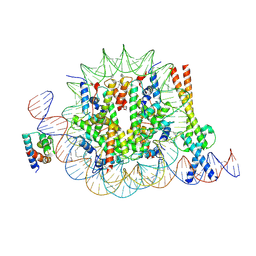 | | OCT4 and MYC-MAX co-bound to a nucleosome | | 分子名称: | DNA (127-MER), Green fluorescent protein,POU domain, class 5, ... | | 著者 | Michael, A.K, Stoos, L, Kempf, G, Cavadini, S, Thoma, N. | | 登録日 | 2023-04-21 | | 公開日 | 2023-05-24 | | 最終更新日 | 2023-07-26 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Cooperation between bHLH transcription factors and histones for DNA access.
Nature, 619, 2023
|
|
8OTT
 
 | | MYC-MAX bound to a nucleosome at SHL+5.8 | | 分子名称: | DNA (144-MER), Histone H2A type 1-B/E, Histone H2A type 1-K, ... | | 著者 | Stoos, L, Michael, A.K, Kempf, G, Kater, L, Cavadini, S, Thoma, N. | | 登録日 | 2023-04-21 | | 公開日 | 2023-05-24 | | 最終更新日 | 2023-09-06 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Cooperation between bHLH transcription factors and histones for DNA access.
Nature, 619, 2023
|
|
8OSK
 
 | | Cryo-EM structure of CLOCK-BMAL1 bound to a nucleosomal E-box at position SHL+5.8 (composite map) | | 分子名称: | Basic helix-loop-helix ARNT-like protein 1, Circadian locomoter output cycles protein kaput, DNA (124-MER), ... | | 著者 | Stoos, L, Michael, A.K, Kempf, G, Cavadini, S, Thoma, N.H. | | 登録日 | 2023-04-19 | | 公開日 | 2023-05-24 | | 最終更新日 | 2023-07-26 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Cooperation between bHLH transcription factors and histones for DNA access.
Nature, 619, 2023
|
|
8OSJ
 
 | | Cryo-EM structure of CLOCK-BMAL1 bound to a nucleosomal E-box at position SHL-6.2 (DNA conformation 1) | | 分子名称: | Basic helix-loop-helix ARNT-like protein 1, Circadian locomoter output cycles protein kaput, DNA (124-MER), ... | | 著者 | Michael, A.K, Stoos, L, Kempf, G, Cavadini, S, Thoma, N.H. | | 登録日 | 2023-04-19 | | 公開日 | 2023-05-24 | | 最終更新日 | 2023-07-26 | | 実験手法 | ELECTRON MICROSCOPY (6.2 Å) | | 主引用文献 | Cooperation between bHLH transcription factors and histones for DNA access.
Nature, 619, 2023
|
|
7Q3E
 
 | | Structure of the mouse CPLANE-RSG1 complex | | 分子名称: | Ciliogenesis and planar polarity effector 2, GUANOSINE-5'-TRIPHOSPHATE, Protein fuzzy homolog, ... | | 著者 | Langousis, G, Cavadini, S, Kempf, G, Matthias, P. | | 登録日 | 2021-10-27 | | 公開日 | 2022-04-06 | | 最終更新日 | 2022-04-27 | | 実験手法 | ELECTRON MICROSCOPY (3.35 Å) | | 主引用文献 | Structure of the ciliogenesis-associated CPLANE complex.
Sci Adv, 8, 2022
|
|
7Q3D
 
 | | Structure of the human CPLANE complex | | 分子名称: | Protein fuzzy homolog, Protein inturned, WD repeat-containing and planar cell polarity effector protein fritz homolog | | 著者 | Langousis, G, Cavadini, S, Kempf, G, Matthias, P. | | 登録日 | 2021-10-27 | | 公開日 | 2022-04-06 | | 最終更新日 | 2022-04-27 | | 実験手法 | ELECTRON MICROSCOPY (3.35 Å) | | 主引用文献 | Structure of the ciliogenesis-associated CPLANE complex.
Sci Adv, 8, 2022
|
|
8AJM
 
 | | Structure of human DDB1-DCAF12 in complex with the C-terminus of CCT5 | | 分子名称: | DDB1- and CUL4-associated factor 12, DNA damage-binding protein 1, T-complex protein 1 subunit epsilon | | 著者 | Pla-Prats, C, Cavadini, S, Kempf, G, Thoma, N.H. | | 登録日 | 2022-07-28 | | 公開日 | 2022-11-09 | | 最終更新日 | 2023-05-24 | | 実験手法 | ELECTRON MICROSCOPY (2.83 Å) | | 主引用文献 | Recognition of the CCT5 di-Glu degron by CRL4 DCAF12 is dependent on TRiC assembly.
Embo J., 42, 2023
|
|
8AJN
 
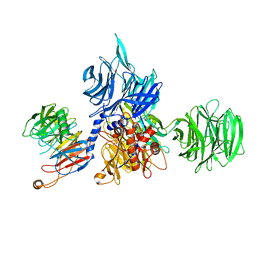 | | Structure of the human DDB1-DCAF12 complex | | 分子名称: | DDB1- and CUL4-associated factor 12, DNA damage-binding protein 1 | | 著者 | Pla-Prats, C, Cavadini, S, Kempf, G, Thoma, N.H. | | 登録日 | 2022-07-28 | | 公開日 | 2022-11-09 | | 最終更新日 | 2023-05-24 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Recognition of the CCT5 di-Glu degron by CRL4 DCAF12 is dependent on TRiC assembly.
Embo J., 42, 2023
|
|
8AJO
 
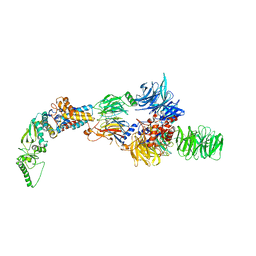 | | Negative-stain electron microscopy structure of DDB1-DCAF12-CCT5 | | 分子名称: | DDB1- and CUL4-associated factor 12, DNA damage-binding protein 1, T-complex protein 1 subunit epsilon | | 著者 | Pla-Prats, C, Cavadini, S, Kempf, G, Thoma, N.H. | | 登録日 | 2022-07-28 | | 公開日 | 2022-11-09 | | 最終更新日 | 2023-05-24 | | 実験手法 | ELECTRON MICROSCOPY (30.6 Å) | | 主引用文献 | Recognition of the CCT5 di-Glu degron by CRL4 DCAF12 is dependent on TRiC assembly.
Embo J., 42, 2023
|
|
6YOV
 
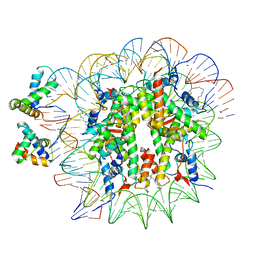 | | OCT4-SOX2-bound nucleosome - SHL+6 | | 分子名称: | DNA (142-MER), Green fluorescent protein,POU domain, class 5, ... | | 著者 | Michael, A.K, Kempf, G, Cavadini, S, Bunker, R.D, Thoma, N.H. | | 登録日 | 2020-04-15 | | 公開日 | 2020-05-06 | | 最終更新日 | 2020-07-08 | | 実験手法 | ELECTRON MICROSCOPY (3.42 Å) | | 主引用文献 | Mechanisms of OCT4-SOX2 motif readout on nucleosomes.
Science, 368, 2020
|
|
6Y5D
 
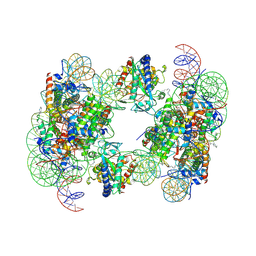 | | Structure of human cGAS (K394E) bound to the nucleosome | | 分子名称: | Cyclic GMP-AMP synthase, DNA (153-MER), Histone H2A type 2-A, ... | | 著者 | Pathare, G.R, Cavadini, S, Kempf, G, Thoma, N.H. | | 登録日 | 2020-02-25 | | 公開日 | 2020-09-23 | | 最終更新日 | 2020-12-09 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structural mechanism of cGAS inhibition by the nucleosome.
Nature, 587, 2020
|
|
7O2W
 
 | | Structure of the C9orf72-SMCR8 complex | | 分子名称: | Guanine nucleotide exchange protein SMCR8,Guanine nucleotide exchange protein SMCR8,Maltose/maltodextrin-binding periplasmic protein, Ubiquitin-like protein SMT3,Guanine nucleotide exchange C9orf72 | | 著者 | Noerpel, J, Cavadini, S, Schenk, A.D, Graff-Meyer, A, Chao, J, Bhaskar, V. | | 登録日 | 2021-03-31 | | 公開日 | 2021-07-21 | | 最終更新日 | 2022-02-02 | | 実験手法 | ELECTRON MICROSCOPY | | 主引用文献 | Structure of the human C9orf72-SMCR8 complex reveals a multivalent protein interaction architecture.
Plos Biol., 19, 2021
|
|
7OKQ
 
 | | Cryo-EM Structure of the DDB1-DCAF1-CUL4A-RBX1 Complex | | 分子名称: | Cullin-4A, DDB1- and CUL4-associated factor 1, DNA damage-binding protein 1, ... | | 著者 | Mohamed, W.I, Schenk, A.D, Kempf, G, Cavadini, S, Thoma, N.H. | | 登録日 | 2021-05-18 | | 公開日 | 2021-10-13 | | 最終更新日 | 2021-12-01 | | 実験手法 | ELECTRON MICROSCOPY (8.4 Å) | | 主引用文献 | The CRL4 DCAF1 cullin-RING ubiquitin ligase is activated following a switch in oligomerization state.
Embo J., 40, 2021
|
|
6Y5E
 
 | | Structure of human cGAS (K394E) bound to the nucleosome (focused refinement of cGAS-NCP subcomplex) | | 分子名称: | Cyclic GMP-AMP synthase, DNA (153-MER), Histone H2A type 2-C, ... | | 著者 | Pathare, G.R, Cavadini, S, Kempf, G, Thoma, N.H. | | 登録日 | 2020-02-25 | | 公開日 | 2020-09-23 | | 最終更新日 | 2020-12-09 | | 実験手法 | ELECTRON MICROSCOPY (3.15 Å) | | 主引用文献 | Structural mechanism of cGAS inhibition by the nucleosome.
Nature, 587, 2020
|
|
6R8Z
 
 | | Cryo-EM structure of NCP_THF2(-1)-UV-DDB | | 分子名称: | DNA damage-binding protein 1, DNA damage-binding protein 2, Histone H2A type 1-B/E, ... | | 著者 | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | 登録日 | 2019-04-02 | | 公開日 | 2019-06-12 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
6R94
 
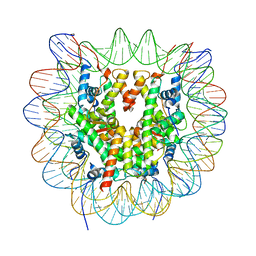 | | Cryo-EM structure of NCP_THF2(-3) | | 分子名称: | Histone H2A type 1-B/E, Histone H2B type 1-J, Histone H3.1, ... | | 著者 | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | 登録日 | 2019-04-02 | | 公開日 | 2019-06-12 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
6R91
 
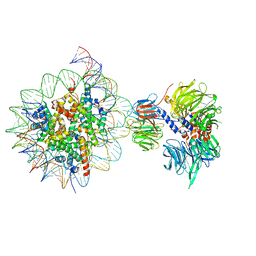 | | Cryo-EM structure of NCP_THF2(-3)-UV-DDB | | 分子名称: | DNA damage-binding protein 1, DNA damage-binding protein 2, Histone H2A type 1-B/E, ... | | 著者 | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | 登録日 | 2019-04-02 | | 公開日 | 2019-06-12 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
6R8Y
 
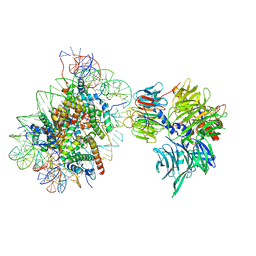 | | Cryo-EM structure of NCP-6-4PP(-1)-UV-DDB | | 分子名称: | DNA damage-binding protein 1, DNA damage-binding protein 2, Histone H2A type 1-B/E, ... | | 著者 | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | 登録日 | 2019-04-02 | | 公開日 | 2019-06-12 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
6R93
 
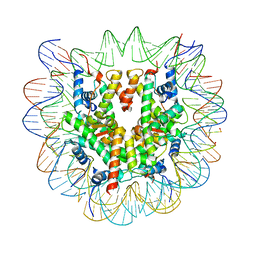 | | Cryo-EM structure of NCP-6-4PP | | 分子名称: | Histone H2A type 1-B/E, Histone H2B type 1-J, Histone H3.1, ... | | 著者 | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | 登録日 | 2019-04-02 | | 公開日 | 2019-06-12 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
6R92
 
 | | Cryo-EM structure of NCP-THF2(+1)-UV-DDB class B | | 分子名称: | DNA damage-binding protein 1,DNA damage-binding protein 1, DNA damage-binding protein 2, Histone H2A type 1-B/E, ... | | 著者 | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | 登録日 | 2019-04-02 | | 公開日 | 2019-06-12 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
6R90
 
 | | Cryo-EM structure of NCP-THF2(+1)-UV-DDB class A | | 分子名称: | DNA damage-binding protein 1, DNA damage-binding protein 2, Histone H2A type 1-B/E, ... | | 著者 | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | 登録日 | 2019-04-02 | | 公開日 | 2019-06-12 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
