3I4H
 
 | | Crystal structure of Cas6 in Pyrococcus furiosus | | Descriptor: | endoribonuclease | | Authors: | Carte, J, Wang, R, Li, H, Terns, R.M, Terns, M.P. | | Deposit date: | 2009-07-01 | | Release date: | 2009-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes
Genes Dev., 22, 2008
|
|
4TVX
 
 | | Crystal structure of the E. coli CRISPR RNA-guided surveillance complex, Cascade | | Descriptor: | CRISPR system Cascade subunit CasA, CRISPR system Cascade subunit CasB, CRISPR system Cascade subunit CasC, ... | | Authors: | Jackson, R.N, Golden, S.M, Carter, J, Wiedenheft, B. | | Deposit date: | 2014-06-28 | | Release date: | 2014-08-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Structural biology. Crystal structure of the CRISPR RNA-guided surveillance complex from Escherichia coli.
Science, 345, 2014
|
|
8CXC
 
 | | Novel Anti-Mesothelin Antibodies Enable Crystallography of the Intact Mesothelin Ectodo- main and Engineering of Potent, T cell-engaging Bispecific Therapeutics | | Descriptor: | 3F2 Antibody heavy chain, 3F2 Antibody light chain, Mesothelin, ... | | Authors: | Bandaranayake, A.D, Rupert, P.B, Lin, I, Pilat, K, Ruff, R.O, Friend, D.J, Chan, M.K, Clarke, M, Carter, J, Meshinchi, S, Mehlin, C, Olson, J.M, Strong, R.K, Correnti, C.E. | | Deposit date: | 2022-05-20 | | Release date: | 2023-06-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (4.31 Å) | | Cite: | Novel mesothelin antibodies enable crystallography of the intact mesothelin ectodomain and engineering of potent, T cell-engaging bispecific therapeutics
Front. Drug Discov., 3, 2023
|
|
8CYH
 
 | | Novel Anti-Mesothelin Antibodies Enable Crystallography of the Intact Mesothelin Ectodo- main and Engineering of Potent, T cell-engaging Bispecific Therapeutics | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, A12 antibody heavy chain, A12 antibody light chain, ... | | Authors: | Bandaranayake, A.D, Rupert, P.B, Lin, I, Pilat, K, Ruff, R.O, Friend, D.J, Chan, M.K, Clarke, M, Carter, J, Meshinchi, S, Mehlin, C, Olson, J.M, Strong, R.K, Correnti, C.E. | | Deposit date: | 2022-05-23 | | Release date: | 2023-06-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | Novel mesothelin antibodies enable crystallography of the intact mesothelin ectodomain and engineering of potent, T cell-engaging bispecific therapeutics
Front. Drug Discov., 3, 2023
|
|
8CZ8
 
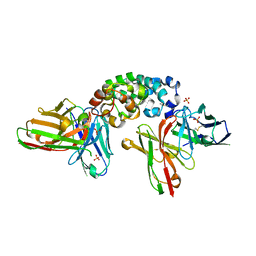 | | Novel Anti-Mesothelin Antibodies Enable Crystallography of the Intact Mesothelin Ectodo- main and Engineering of Potent, T cell-engaging Bispecific Therapeutics | | Descriptor: | Mesothelin, cleaved form, SULFATE ION, ... | | Authors: | Bandaranayake, A.D, Rupert, P.B, Lin, I, Pilat, K, Ruff, R.O, Friend, D.J, Chan, M.K, Clarke, M, Carter, J, Meshinchi, S, Mehlin, C, Olson, J.M, Strong, R.K, Correnti, C.E. | | Deposit date: | 2022-05-24 | | Release date: | 2023-06-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Novel mesothelin antibodies enable crystallography of the intact mesothelin ectodomain and engineering of potent, T cell-engaging bispecific therapeutics
Front. Drug Discov., 3, 2023
|
|
5UZ9
 
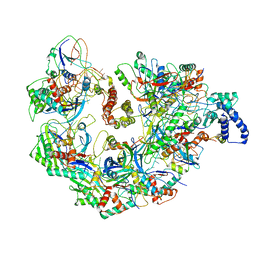 | | Cryo EM structure of anti-CRISPRs, AcrF1 and AcrF2, bound to type I-F crRNA-guided CRISPR surveillance complex | | Descriptor: | Anti-CRISPR protein 30, Anti-CRISPR protein Acr30-35, CRISPR RNA (60-MER), ... | | Authors: | Chowdhury, S, Carter, J, Rollins, M.F, Jackson, R.N, Hoffmann, C, Nosaka, L, Bondy-Denomy, J, Maxwell, K.L, Davidson, A.R, Fischer, E.R, Lander, G.C, Wiedenheft, B. | | Deposit date: | 2017-02-25 | | Release date: | 2017-04-26 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure Reveals Mechanisms of Viral Suppressors that Intercept a CRISPR RNA-Guided Surveillance Complex.
Cell, 169, 2017
|
|
5CD4
 
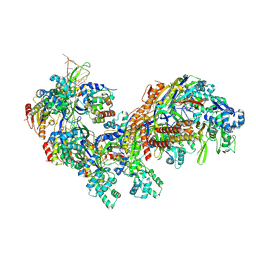 | | The Type IE CRISPR Cascade complex from E. coli, with two assemblies in the asymmetric unit arranged back-to-back | | Descriptor: | CRISPR system Cascade subunit CasA, CRISPR system Cascade subunit CasB, CRISPR system Cascade subunit CasC, ... | | Authors: | Jackson, R.N, Golden, S.M, Carter, J, Wiedenheft, B. | | Deposit date: | 2015-07-03 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Mechanism of CRISPR-RNA guided recognition of DNA targets in Escherichia coli.
Nucleic Acids Res., 43, 2015
|
|
6NE0
 
 | | Structure of double-stranded target DNA engaged Csy complex from Pseudomonas aeruginosa (PA-14) | | Descriptor: | CRISPR RNA (60-MER), CRISPR target DNA (44-MER), CRISPR-associated endonuclease Cas6/Csy4, ... | | Authors: | Chowdhury, S, Rollins, M.F, Carter, J, Golden, S.M, Miettinen, H.M, Santiago-Frangos, A, Faith, D, Lawrence, M.C, Wiedenheft, B, Lander, G.C. | | Deposit date: | 2018-12-15 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure Reveals a Mechanism of CRISPR-RNA-Guided Nuclease Recruitment and Anti-CRISPR Viral Mimicry.
Mol. Cell, 74, 2019
|
|
3NTG
 
 | | Crystal structure of COX-2 with selective compound 23d-(R) | | Descriptor: | (2R)-6,8-dichloro-7-(2-methylpropoxy)-2-(trifluoromethyl)-2H-chromene-3-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, J.L, Limburg, D, Graneto, M.J, Carter, J.C, Talley, J.J, Kiefer, J.R. | | Deposit date: | 2010-07-04 | | Release date: | 2010-10-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The novel benzopyran class of selective cyclooxygenase-2 inhibitors. Part 2: The second clinical candidate having a shorter and favorable human half-life.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
8P0D
 
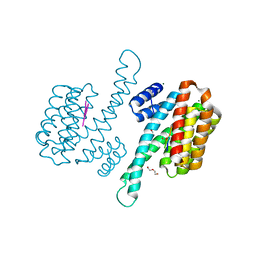 | | Human 14-3-3 sigma in complex with human MDM2 peptide | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Roversi, P, Ward, J, Doveston, R, Kwon, H, Romartinez Alonso, B. | | Deposit date: | 2023-05-10 | | Release date: | 2024-02-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Characterizing the protein-protein interaction between MDM2 and 14-3-3 sigma ; proof of concept for small molecule stabilization.
J.Biol.Chem., 300, 2024
|
|
6NMV
 
 | | Non-Blocking Fab 218 anti-SIRP-alpha antibody in complex with SIRP-alpha Variant 1 | | Descriptor: | Fab 218 anti-SIRP-alpha antibody Variable Heavy Chain, Fab 218 anti-SIRP-alpha antibody Variable Light Chain, Tyrosine-protein phosphatase non-receptor type substrate 1 | | Authors: | Wibowo, A.S, Carter, J.J, Sim, J. | | Deposit date: | 2019-01-11 | | Release date: | 2019-08-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Discovery of high affinity, pan-allelic, and pan-mammalian reactive antibodies against the myeloid checkpoint receptor SIRP alpha.
Mabs, 11, 2019
|
|
6NMT
 
 | | Non-Blocking Fab 3 anti-SIRP-alpha antibody in complex with SIRP-alpha Variant 1 | | Descriptor: | Fab 3 anti-SIRP-alpha antibody Variable Heavy Chain, Fab 3 anti-SIRP-alpha antibody Variable Light Chain, Tyrosine-protein phosphatase non-receptor type substrate 1 | | Authors: | Wibowo, A.S, Carter, J.J, Sim, J. | | Deposit date: | 2019-01-11 | | Release date: | 2019-08-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery of high affinity, pan-allelic, and pan-mammalian reactive antibodies against the myeloid checkpoint receptor SIRP alpha.
Mabs, 11, 2019
|
|
6NMS
 
 | | Blocking Fab 136 anti-SIRP-alpha antibody in complex with SIRP-alpha Variant 1 | | Descriptor: | Fab 136 anti-SIRP-alpha antibody Variable Heavy Chain, Fab 136 anti-SIRP-alpha antibody Variable Light Chain, Tyrosine-protein phosphatase non-receptor type substrate 1 | | Authors: | Wibowo, A.S, Carter, J.J, Sim, J. | | Deposit date: | 2019-01-11 | | Release date: | 2019-08-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Discovery of high affinity, pan-allelic, and pan-mammalian reactive antibodies against the myeloid checkpoint receptor SIRP alpha.
Mabs, 11, 2019
|
|
6NMU
 
 | | Kick-Off Fab 115 anti-SIRP-alpha antibody in complex with SIRP-alpha Variant 1 | | Descriptor: | Fab 115 anti-SIRP-alpha antibody Variable Heavy Chain, Fab 115 anti-SIRP-alpha antibody Variable Light Chain, Tyrosine-protein phosphatase non-receptor type substrate 1 | | Authors: | Wibowo, A.S, Carter, J.J, Sim, J. | | Deposit date: | 2019-01-11 | | Release date: | 2019-08-07 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery of high affinity, pan-allelic, and pan-mammalian reactive antibodies against the myeloid checkpoint receptor SIRP alpha.
Mabs, 11, 2019
|
|
6NMR
 
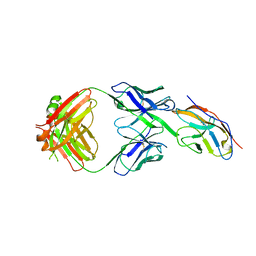 | | Blocking Fab 119 anti-SIRP-alpha antibody in complex with SIRP-alpha Variant 1 | | Descriptor: | Fab 119 anti-SIRP-alpha antibody Variable Heavy Chain, Fab 119 anti-SIRP-alpha antibody Variable Light Chain, Tyrosine-protein phosphatase non-receptor type substrate 1 | | Authors: | Wibowo, A.S, Carter, J.J, Sim, J. | | Deposit date: | 2019-01-11 | | Release date: | 2019-08-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Discovery of high affinity, pan-allelic, and pan-mammalian reactive antibodies against the myeloid checkpoint receptor SIRP alpha.
Mabs, 11, 2019
|
|
3MQE
 
 | | Structure of SC-75416 bound at the COX-2 active site | | Descriptor: | (2S)-7-tert-butyl-6-chloro-2-(trifluoromethyl)-2H-chromene-3-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, J.L, Limburg, D, Graneto, M.J, Springer, J, Rogier, J, Kiefer, J.R. | | Deposit date: | 2010-04-28 | | Release date: | 2010-10-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The novel benzopyran class of selective cyclooxygenase-2 inhibitors. Part 2: The second clinical candidate having a shorter and favorable human half-life.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
6P8Z
 
 | | Crystal structure of human KRAS G12C covalently bound to an acryloylazetidine acetamide inhibitor | | Descriptor: | 2-[5-chloro-2-cyclopropyl-3-(5-methoxy-3,4-dihydroisoquinoline-2(1H)-carbonyl)-7-methyl-1H-indol-1-yl]-N-(1-propanoylazetidin-3-yl)acetamide, CALCIUM ION, GTPase KRas, ... | | Authors: | Mohr, C. | | Deposit date: | 2019-06-08 | | Release date: | 2019-08-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery ofN-(1-Acryloylazetidin-3-yl)-2-(1H-indol-1-yl)acetamides as Covalent Inhibitors of KRASG12C.
Acs Med.Chem.Lett., 10, 2019
|
|
6P8W
 
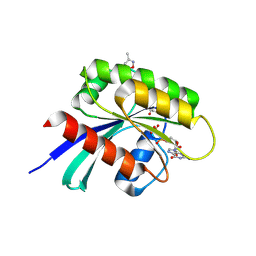 | |
3LN0
 
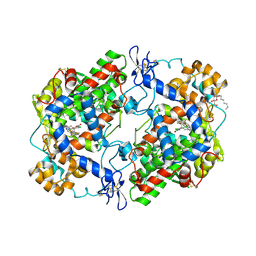 | | Structure of compound 5c-S bound at the active site of COX-2 | | Descriptor: | (2S)-6,8-dichloro-2-(trifluoromethyl)-2H-chromene-3-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kiefer, J.R, Kurumbail, R.G, Stallings, W.C, Pawlitz, J.L. | | Deposit date: | 2010-02-01 | | Release date: | 2010-10-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The novel benzopyran class of selective cyclooxygenase-2 inhibitors. Part 2: The second clinical candidate having a shorter and favorable human half-life.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
6P8Y
 
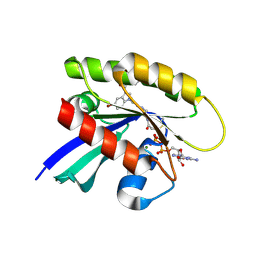 | |
6P8X
 
 | |
3LN1
 
 | | Structure of celecoxib bound at the COX-2 active site | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[5-(4-METHYLPHENYL)-3-(TRIFLUOROMETHYL)-1H-PYRAZOL-1-YL]BENZENESULFONAMIDE, ... | | Authors: | Kiefer, J.R, Kurumbail, R.G, Stallings, W.C, Pawlitz, J.L. | | Deposit date: | 2010-02-01 | | Release date: | 2010-10-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The novel benzopyran class of selective cyclooxygenase-2 inhibitors. Part 2: The second clinical candidate having a shorter and favorable human half-life.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
5KAE
 
 | |
3S2A
 
 | | Crystal structure of PI3K-gamma in complex with a quinoline inhibitor | | Descriptor: | N-{2-chloro-5-[4-(morpholin-4-yl)quinolin-6-yl]pyridin-3-yl}-4-fluorobenzenesulfonamide, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Whittington, D.A, Tang, J, Yakowec, P. | | Deposit date: | 2011-05-16 | | Release date: | 2011-06-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Phospshoinositide 3-Kinase (PI3K)/Mammalian Target of Rapamycin (mTOR) Dual Inhibitors: Discovery and Structure-Activity Relationships of a Series of Quinoline and Quinoxaline Derivatives.
J.Med.Chem., 54, 2011
|
|
7MU4
 
 | | Crystal Structure of HPV L1-directed D24.M01Fab | | Descriptor: | D24.M01 Fab Heavy Chain, D24.M01 Fab Light Chain, DI(HYDROXYETHYL)ETHER | | Authors: | Singh, S, Pancera, M. | | Deposit date: | 2021-05-14 | | Release date: | 2022-05-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural Characterisation of Immune Responses to HPV Vaccination
To Be Published
|
|
