6FOE
 
 | | BaxB01 Fab fragment | | Descriptor: | Fab BaxB01 heavy chain, Fab BaxB01 light chain | | Authors: | Hollerweger, J, Schinagl, A, Kerschbaumer, R.J, Scheiflinger, F, Thiele, M, Goettig, P, Brandstetter, H. | | Deposit date: | 2018-02-07 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Role of the Cysteine 81 Residue of Macrophage Migration Inhibitory Factor as a Molecular Redox Switch.
Biochemistry, 57, 2018
|
|
6YSA
 
 | | Crystal structure of Arabidopsis thaliana legumain isoform beta in zymogen state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, SULFATE ION, ... | | Authors: | Dall, E, Zauner, F.B, Brandstetter, H. | | Deposit date: | 2020-04-21 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural and functional studies ofArabidopsis thalianalegumain beta reveal isoform specific mechanisms of activation and substrate recognition.
J.Biol.Chem., 295, 2020
|
|
5O7E
 
 | |
5NZC
 
 | |
5NZB
 
 | |
5OBT
 
 | |
7QXZ
 
 | | X-ray structure of furin in complex with the dichlorophenylpyridine-based inhibitor 5 | | Descriptor: | 2-[(3S)-1-[[2-[3,5-bis(chloranyl)phenyl]-6-[2-(4-methylpiperazin-4-ium-1-yl)pyrimidin-5-yl]oxy-pyridin-4-yl]methyl]pyrrolidin-1-ium-3-yl]oxyethanoic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Dahms, S.O, Brandstetter, H, Pautsch, A. | | Deposit date: | 2022-01-27 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Dichlorophenylpyridine-Based Molecules Inhibit Furin through an Induced-Fit Mechanism.
Acs Chem.Biol., 17, 2022
|
|
7QY1
 
 | | X-ray structure of furin in complex with the dichlorophenylpyridine-based inhibitor 4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[4-[5-[4-[[4-(acetamidomethyl)piperidin-1-ium-1-yl]methyl]-6-[3,5-bis(chloranyl)phenyl]pyridin-2-yl]oxypyridin-2-yl]piperazin-1-ium-1-yl]propanoate, CALCIUM ION, ... | | Authors: | Dahms, S.O, Brandstetter, H, Pautsch, A. | | Deposit date: | 2022-01-27 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Dichlorophenylpyridine-Based Molecules Inhibit Furin through an Induced-Fit Mechanism.
Acs Chem.Biol., 17, 2022
|
|
7QY0
 
 | | X-ray structure of furin in complex with the dichlorophenylpyridine-based inhibitor 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Dahms, S.O, Brandstetter, H, Pautsch, A. | | Deposit date: | 2022-01-27 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Dichlorophenylpyridine-Based Molecules Inhibit Furin through an Induced-Fit Mechanism.
Acs Chem.Biol., 17, 2022
|
|
7QY2
 
 | | X-ray structure of furin in complex with the dichlorophenylpyridine-based inhibitor 2 | | Descriptor: | (2R)-4-[4-[5-[4-[[4-(acetamidomethyl)piperidin-1-ium-1-yl]methyl]-6-[3,5-bis(chloranyl)phenyl]pyridin-2-yl]oxypyrimidin-2-yl]piperazin-1-ium-1-yl]-2-methyl-butanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Dahms, S.O, Brandstetter, H, Pautsch, A. | | Deposit date: | 2022-01-27 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Dichlorophenylpyridine-Based Molecules Inhibit Furin through an Induced-Fit Mechanism.
Acs Chem.Biol., 17, 2022
|
|
7QXY
 
 | | X-ray structure of furin in complex with the dichlorophenylpyridine-based inhibitor 3 | | Descriptor: | 3-[4-[5-[4-[[4-(acetamidomethyl)piperidin-1-ium-1-yl]methyl]-6-[3,5-bis(chloranyl)phenyl]pyridin-2-yl]oxypyrimidin-2-yl]piperazin-1-ium-1-yl]propanoate, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Dahms, S.O, Brandstetter, H, Pautsch, A. | | Deposit date: | 2022-01-27 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.478 Å) | | Cite: | Dichlorophenylpyridine-Based Molecules Inhibit Furin through an Induced-Fit Mechanism.
Acs Chem.Biol., 17, 2022
|
|
7O1U
 
 | |
7O22
 
 | |
7O20
 
 | |
7O1W
 
 | |
7O1Y
 
 | |
7O50
 
 | | Crystal structure of human legumain in complex with Gly-Ser-Asn peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLY-SER-ASN, ... | | Authors: | Dall, E, Brandstetter, H. | | Deposit date: | 2021-04-07 | | Release date: | 2021-09-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Peptide Ligase Activity of Human Legumain Depends on Fold Stabilization and Balanced Substrate Affinities.
Acs Catalysis, 11, 2021
|
|
1Z1W
 
 | | Crystal structures of the tricorn interacting facor F3 from Thermoplasma acidophilum, a zinc aminopeptidase in three different conformations | | Descriptor: | SULFATE ION, Tricorn protease interacting factor F3, ZINC ION | | Authors: | Kyrieleis, O.J.P, Goettig, P, Kiefersauer, R, Huber, R, Brandstetter, H. | | Deposit date: | 2005-03-07 | | Release date: | 2005-05-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structures of the Tricorn Interacting Factor F3 from Thermoplasma acidophilum, a Zinc Aminopeptidase in Three Different Conformations
J.MOL.BIOL., 394, 2005
|
|
1Z5H
 
 | | Crystal structures of the Tricorn interacting Factor F3 from Thermoplasma acidophilum | | Descriptor: | SULFATE ION, Tricorn protease interacting factor F3, ZINC ION | | Authors: | Kyrieleis, O.J.P, Goettig, P, Kiefersauer, R, Huber, R, Brandstetter, H. | | Deposit date: | 2005-03-18 | | Release date: | 2005-06-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of the Tricorn Interacting Factor F3 from Thermoplasma acidophilum, a Zinc Aminopeptidase in Three Different Conformations
J.MOL.BIOL., 349, 2005
|
|
1ZRZ
 
 | | Crystal Structure of the Catalytic Domain of Atypical Protein Kinase C-iota | | Descriptor: | 3-{1-[3-(DIMETHYLAMINO)PROPYL]-1H-INDOL-3-YL}-4-(1H-INDOL-3-YL)-1H-PYRROLE-2,5-DIONE, Protein kinase C, iota | | Authors: | Messerschmidt, A, Macieira, S, Velarde, M, Baedeker, M, Benda, C, Jestel, A, Brandstetter, H, Neuefeind, T, Blaesse, M, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-05-23 | | Release date: | 2005-09-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of the Catalytic Domain of Human Atypical Protein Kinase C-iota Reveals Interaction Mode of Phosphorylation Site in Turn Motif
J.Mol.Biol., 352, 2005
|
|
5LUA
 
 | | Crystal structure of human legumain (AEP) in complex with compound 11b | | Descriptor: | 2,4-di(morpholin-4-yl)aniline, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dall, E, Ye, K, Brandstetter, H. | | Deposit date: | 2016-09-08 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibition of delta-secretase improves cognitive functions in mouse models of Alzheimer's disease.
Nat Commun, 8, 2017
|
|
5LUB
 
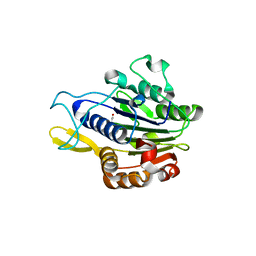 | | Crystal structure of human legumain (AEP) in complex with compound 11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 7-(morpholin-4-yl)-2,1,3-benzoxadiazol-4-amine, ... | | Authors: | Dall, E, Ye, K, Brandstetter, H. | | Deposit date: | 2016-09-08 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibition of delta-secretase improves cognitive functions in mouse models of Alzheimer's disease.
Nat Commun, 8, 2017
|
|
5LU8
 
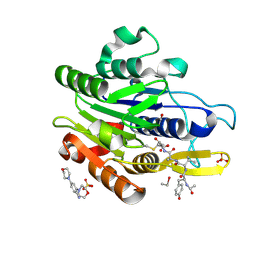 | | CRYSTAL STRUCTURE OF YVAD-CMK BOUND HUMAN LEGUMAIN (AEP) IN COMPLEX WITH COMPOUND 11B | | Descriptor: | 2,4-di(morpholin-4-yl)aniline, 2-acetamido-2-deoxy-beta-D-glucopyranose, AC-TYR-VAL-ALA-ASP-CHLOROMETHYLKETONE, ... | | Authors: | Dall, E, Ye, K, Brandstetter, H. | | Deposit date: | 2016-09-08 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Inhibition of delta-secretase improves cognitive functions in mouse models of Alzheimer's disease.
Nat Commun, 8, 2017
|
|
5LU9
 
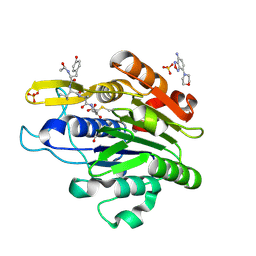 | | Crystal structure of YVAD-cmk bound human legumain (AEP) in complex with compound 11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 7-(morpholin-4-yl)-2,1,3-benzoxadiazol-4-amine, AC-TYR-VAL-ALA-ASP-CHLOROMETHYLKETONE, ... | | Authors: | Dall, E, Ye, K, Brandstetter, H. | | Deposit date: | 2016-09-08 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Inhibition of delta-secretase improves cognitive functions in mouse models of Alzheimer's disease.
Nat Commun, 8, 2017
|
|
7ZBV
 
 | |
