1TVF
 
 | | Crystal Structure of penicillin-binding protein 4 (PBP4) from Staphylococcus aureus | | 分子名称: | SULFATE ION, UNKNOWN LIGAND, penicillin binding protein 4 | | 著者 | Rajashankar, K.R, Ray, S.S, Bonanno, J.B, Pinho, M, Tomasz, A, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2004-06-29 | | 公開日 | 2004-07-06 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal Structure of penicillin-binding protein 4 (PBP4) from Staphylococcus aureus
To be Published
|
|
1JSX
 
 | |
1K47
 
 | |
1KU9
 
 | | X-ray Structure of a Methanococcus jannaschii DNA-Binding Protein: Implications for Antibiotic Resistance in Staphylococcus aureus | | 分子名称: | hypothetical protein MJ223 | | 著者 | Ray, S.S, Bonanno, J.B, Chen, H, de Lencastre, H, Wu, S, Tomasz, A, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2002-01-21 | | 公開日 | 2002-12-25 | | 最終更新日 | 2021-02-03 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | X-ray structure of an M. jannaschii DNA-binding protein: implications for antibiotic resistance in S.
aureus
Proteins, 50, 2002
|
|
6OI0
 
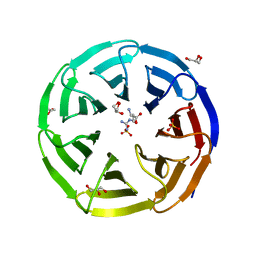 | | Crystal structure of human WDR5 in complex with L-arginine | | 分子名称: | ARGININE, GLYCEROL, SULFATE ION, ... | | 著者 | Lorton, B.M, Harijan, R.K, Burgos, E, Bonanno, J.B, Almo, S.C, Shechter, D. | | 登録日 | 2019-04-08 | | 公開日 | 2020-04-01 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | A Binary Arginine Methylation Switch on Histone H3 Arginine 2 Regulates Its Interaction with WDR5.
Biochemistry, 59, 2020
|
|
6OI3
 
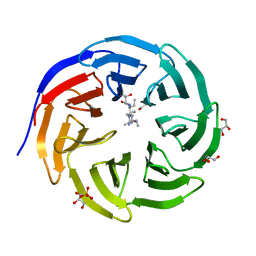 | | Crystal structure of human WDR5 in complex with monomethyl H3R2 peptide | | 分子名称: | GLYCEROL, Monomethyl H3R2 peptide, SULFATE ION, ... | | 著者 | Lorton, B.M, Harijan, R.K, Burgos, E, Bonanno, J.B, Almo, S.C, Shechter, D. | | 登録日 | 2019-04-08 | | 公開日 | 2020-04-01 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | A Binary Arginine Methylation Switch on Histone H3 Arginine 2 Regulates Its Interaction with WDR5.
Biochemistry, 59, 2020
|
|
6OFZ
 
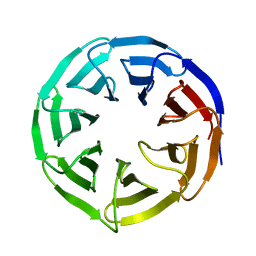 | | Crystal structure of human WDR5 | | 分子名称: | WD repeat-containing protein 5 | | 著者 | Lorton, B.M, Harijan, R.K, Burgos, E, Bonanno, J.B, Almo, S.C, Shechter, D. | | 登録日 | 2019-04-01 | | 公開日 | 2020-04-01 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | A Binary Arginine Methylation Switch on Histone H3 Arginine 2 Regulates Its Interaction with WDR5.
Biochemistry, 59, 2020
|
|
6OI1
 
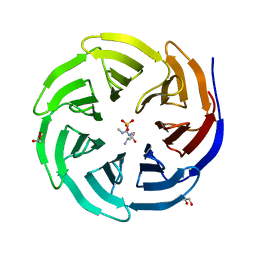 | | Crystal structure of human WDR5 in complex with monomethyl L-arginine | | 分子名称: | (2S)-2-amino-5-[(N-methylcarbamimidoyl)amino]pentanoic acid, GLYCEROL, SULFATE ION, ... | | 著者 | Lorton, B.M, Harijan, R.K, Burgos, E, Bonanno, J.B, Almo, S.C, Shechter, D. | | 登録日 | 2019-04-08 | | 公開日 | 2020-04-01 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | A Binary Arginine Methylation Switch on Histone H3 Arginine 2 Regulates Its Interaction with WDR5.
Biochemistry, 59, 2020
|
|
6DXP
 
 | | The crystal structure of an FMN-dependent NADH-azoreductase from Klebsiella pneumoniae | | 分子名称: | FLAVIN MONONUCLEOTIDE, FMN-dependent NADH-azoreductase | | 著者 | Arcinas, A.J, Ghosh, A, Chamala, S, Bonanno, J.B, Kelly, L, Almo, S.C. | | 登録日 | 2018-06-29 | | 公開日 | 2018-07-25 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.478 Å) | | 主引用文献 | The crystal structure of an FMN-dependent NADH-azoreductase from Klebsiella pneumoniae
To Be Published
|
|
3BGE
 
 | | Crystal structure of the C-terminal fragment of AAA+ATPase from Haemophilus influenzae | | 分子名称: | Predicted ATPase, SULFATE ION | | 著者 | Ramagopal, U.A, Patskovsky, Y, Bonanno, J.B, Meyer, A.J, Toro, R, Freeman, J, Adams, J, Koss, J, Maletic, M, Gheyi, T, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2007-11-26 | | 公開日 | 2008-01-01 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystal structure of the C-terminal fragment of AAA+ATPase from Haemophilus influenzae.
To be Published
|
|
3BH1
 
 | | Crystal structure of protein DIP2346 from Corynebacterium diphtheriae | | 分子名称: | GLYCEROL, UPF0371 protein DIP2346 | | 著者 | Patskovsky, Y, Sridhar, V, Bonanno, J.B, Gilmore, M, Iizuka, M, Groshong, C, Gheyi, T, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2007-11-27 | | 公開日 | 2007-12-11 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.51 Å) | | 主引用文献 | Crystal structure of protein DIP2346 from Corynebacterium diphtheriae.
To be Published
|
|
3BJS
 
 | | Crystal structure of a member of enolase superfamily from Polaromonas sp. JS666 | | 分子名称: | MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme | | 著者 | Patskovsky, Y, Bonanno, J.B, Ozyurt, S, Dickey, M, Sauder, J.M, Reyes, C, Groshong, C, Gheyi, T, Smith, D, Wasserman, S.R, Gerlt, J, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2007-12-04 | | 公開日 | 2007-12-18 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal Structure of a Member of Enolase Superfamily from Polaromonas sp. JS666.
To be Published
|
|
3B5M
 
 | | Crystal structure of conserved uncharacterized protein from Rhodopirellula baltica | | 分子名称: | SULFATE ION, Uncharacterized protein | | 著者 | Patskovsky, Y, Bonanno, J.B, Sridhar, V, Rutter, M, Powell, A, Maletic, M, Rodgers, R, Wasserman, S, Smith, D, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2007-10-26 | | 公開日 | 2007-11-13 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.21 Å) | | 主引用文献 | Crystal Structure of Conserved Uncharacterized Protein from Rhodopirellula baltica.
To be Published
|
|
3C19
 
 | | Crystal structure of protein MK0293 from Methanopyrus kandleri AV19 | | 分子名称: | GLYCEROL, PHOSPHATE ION, Uncharacterized protein MK0293 | | 著者 | Patskovsky, Y, Romero, R, Bonanno, J.B, Malashkevich, V, Dickey, M, Chang, S, Koss, J, Bain, K, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2008-01-22 | | 公開日 | 2008-02-05 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of protein MK0293 from Methanopyrus kandleri AV19.
To be Published
|
|
3BY5
 
 | | Crystal structure of cobalamin biosynthesis protein chiG from Agrobacterium tumefaciens str. C58 | | 分子名称: | Cobalamin biosynthesis protein, SULFATE ION | | 著者 | Patskovsky, Y, Bonanno, J.B, Sojitra, S, Rutter, M, Iizuka, M, Maletic, M, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2008-01-15 | | 公開日 | 2008-01-22 | | 最終更新日 | 2021-02-03 | | 実験手法 | X-RAY DIFFRACTION (2.52 Å) | | 主引用文献 | Crystal structure of cobalamin biosynthesis protein from Agrobacterium tumefaciens str. C58.
To be Published
|
|
3BMA
 
 | | Crystal structure of D-alanyl-lipoteichoic acid synthetase from Streptococcus pneumoniae R6 | | 分子名称: | D-alanyl-lipoteichoic acid synthetase, GLYCEROL, SULFATE ION | | 著者 | Patskovsky, Y, Sridhar, V, Bonanno, J.B, Smith, D, Rutter, M, Iizuka, M, Koss, J, Bain, K, Gheyi, T, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2007-12-12 | | 公開日 | 2007-12-25 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | Crystal Structure of probable D-Alanyl-Lipoteichoic Acid Synthetase from Streptococcus pneumoniae.
To be Published
|
|
5WMW
 
 | | Structural Insights into Substrate and Inhibitor Binding Sites in Human Indoleamine 2,3-Dioxygenase 1 | | 分子名称: | CYANIDE ION, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Lewis-Ballester, A, Yeh, S.R, Pham, K.N, Batabyal, D, Karkashon, S, Bonanno, J.B, Poulos, T.L. | | 登録日 | 2017-07-31 | | 公開日 | 2017-12-06 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.03 Å) | | 主引用文献 | Structural insights into substrate and inhibitor binding sites in human indoleamine 2,3-dioxygenase 1.
Nat Commun, 8, 2017
|
|
5E5M
 
 | | Crystal structure of mouse CTLA-4 in complex with nanobody | | 分子名称: | CTLA-4 nanobody, Cytotoxic T-lymphocyte protein 4, GLYCEROL | | 著者 | Fedorov, A.A, Fedorov, E.V, Samanta, D, Bonanno, J.B, Almo, S.C. | | 登録日 | 2015-10-08 | | 公開日 | 2016-10-12 | | 最終更新日 | 2019-01-30 | | 実験手法 | X-RAY DIFFRACTION (2.182 Å) | | 主引用文献 | Crystal structure of mouse CTLA-4 in complex with nanobody
To Be Published
|
|
5DK6
 
 | | CRYSTAL STRUCTURE OF A 5'-METHYLTHIOADENOSINE/S-ADENOSYLHOMOCYSTEINE (MTA/SAH) NUCLEOSIDASE (MTAN) FROM COLWELLIA PSYCHRERYTHRAEA 34H (CPS_4743, TARGET PSI-029300) IN COMPLEX WITH ADENINE AT 2.27 A RESOLUTION | | 分子名称: | 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase, ADENINE, GLYCINE | | 著者 | Himmel, D.M, Bhosle, R, Toro, R, Ahmed, M, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Seidel, R.D, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2015-09-03 | | 公開日 | 2015-11-04 | | 実験手法 | X-RAY DIFFRACTION (2.27 Å) | | 主引用文献 | CRYSTAL STRUCTURE OF A 5'-METHYLTHIOADENOSINE/S-ADENOSYLHOMOCYSTEINE (MTA/SAH)NUCLEOSIDASE (MTAN) FROM COLWELLIA PSYCHRERYTHRAEA 34H (CPS_4743, TARGET PSI-029300) IN COMPLEX WITH ADENINE AT 2.27 A RESOLUTION
To be published
|
|
5E56
 
 | | Crystal structure of mouse CTLA-4 | | 分子名称: | Cytotoxic T-lymphocyte protein 4, SODIUM ION | | 著者 | Fedorov, A.A, Fedorov, E.V, SAMANTA, D, Bonanno, J.B, Almo, S.C. | | 登録日 | 2015-10-07 | | 公開日 | 2015-10-28 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.504 Å) | | 主引用文献 | Crystal structure of mouse CTLA-4
To Be Published
|
|
5E03
 
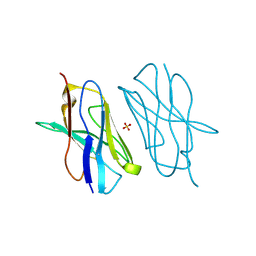 | | Crystal structure of mouse CTLA-4 nanobody | | 分子名称: | CTLA-4 nanobody, SULFATE ION | | 著者 | Fedorov, A.A, Fedorov, E.V, Samanta, D, Bonanno, J.B, Almo, S.C. | | 登録日 | 2015-09-28 | | 公開日 | 2015-10-07 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.685 Å) | | 主引用文献 | Crystal structure of mouse CTLA-4 nanobody
To Be Published
|
|
6C8V
 
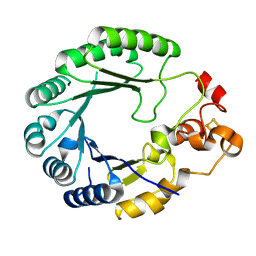 | | X-ray structure of PqqE from Methylobacterium extorquens | | 分子名称: | Coenzyme PQQ synthesis protein E, FE2/S2 (INORGANIC) CLUSTER, IRON/SULFUR CLUSTER | | 著者 | Gizzi, A.S, Grove, T.L, Bonanno, J.B, Almo, S.C. | | 登録日 | 2018-01-25 | | 公開日 | 2018-02-14 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | X-ray and EPR Characterization of the Auxiliary Fe-S Clusters in the Radical SAM Enzyme PqqE.
Biochemistry, 57, 2018
|
|
7MSG
 
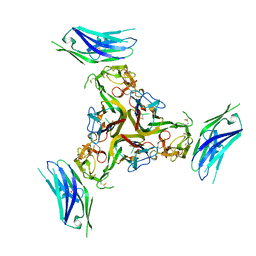 | | The crystal structure of LIGHT in complex with HVEM and CD160 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CD160 antigen, soluble form,Tumor necrosis factor receptor superfamily member 14, ... | | 著者 | Liu, W, Ramagopal, U, Garrett-Thompson, S.C, Fedorov, E, Bonanno, J.B, Almo, S.C. | | 登録日 | 2021-05-11 | | 公開日 | 2021-10-27 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | HVEM structures and mutants reveal distinct functions of binding to LIGHT and BTLA/CD160.
J.Exp.Med., 218, 2021
|
|
7MSJ
 
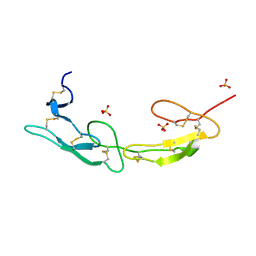 | | The crystal structure of mouse HVEM | | 分子名称: | SULFATE ION, Tumor necrosis factor receptor superfamily member 14 | | 著者 | Liu, W, Ramagopal, U, Garrett-Thompson, S.C, Fedorov, E, Bonanno, J.B, Almo, S.C. | | 登録日 | 2021-05-11 | | 公開日 | 2021-10-27 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | HVEM structures and mutants reveal distinct functions of binding to LIGHT and BTLA/CD160.
J.Exp.Med., 218, 2021
|
|
4NEK
 
 | | Putative enoyl-CoA hydratase/carnithine racemase from Magnetospirillum magneticum AMB-1 | | 分子名称: | DI(HYDROXYETHYL)ETHER, Enoyl-CoA hydratase/carnithine racemase | | 著者 | Tkaczuk, K.L, Cooper, D.R, Geffken, K, Chapman, H.C, Stead, M, Hillerich, B, Ahmed, M, Bonanno, J.B, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2013-10-29 | | 公開日 | 2013-11-27 | | 最終更新日 | 2022-04-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Putative enoyl-CoA hydratase/carnithine racemase from Magnetospirillum magneticum AMB-1
To be Published
|
|
