3UD6
 
 | | Structural analyses of covalent enzyme-substrate analogue complexes reveal strengths and limitations of de novo enzyme design | | Descriptor: | 1-(6-METHOXYNAPHTHALEN-2-YL)BUTANE-1,3-DIONE, RETRO-ALDOLASE, SULFATE ION | | Authors: | Baker, D, Stoddard, B.L, Althoff, E.A, Wang, L, Jiang, L, Moody, J, Bolduc, J, Lassila, J, Hilvert, D. | | Deposit date: | 2011-10-27 | | Release date: | 2011-11-23 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.091 Å) | | Cite: | Structural analyses of covalent enzyme-substrate analog complexes reveal strengths and limitations of de novo enzyme design.
J.Mol.Biol., 415, 2012
|
|
8F6R
 
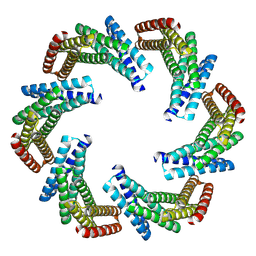 | | CryoEM structure of designed modular protein oligomer C6-79 | | Descriptor: | De novo designed oligomeric protein C6-79 | | Authors: | Redler, R.L, Edman, N.I, Baker, D, Ekiert, D, Bhabha, G. | | Deposit date: | 2022-11-17 | | Release date: | 2023-11-29 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Modulation of FGF pathway signaling and vascular differentiation using designed oligomeric assemblies.
Cell, 187, 2024
|
|
8F6Q
 
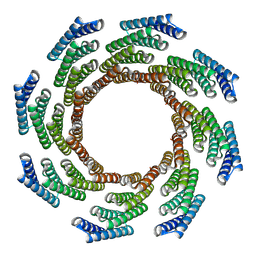 | | CryoEM structure of designed modular protein oligomer C8-71 | | Descriptor: | C8-71 | | Authors: | Redler, R.L, Edman, N.I, Baker, D, Ekiert, D, Bhabha, G. | | Deposit date: | 2022-11-17 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Modulation of FGF pathway signaling and vascular differentiation using designed oligomeric assemblies.
Cell, 187, 2024
|
|
9MRB
 
 | | The designed serine hydrolase known as dad_t1 | | Descriptor: | TETRAETHYLENE GLYCOL, dad_t1 | | Authors: | Pellock, S.J, Lauko, A, Bera, A, Baker, D. | | Deposit date: | 2025-01-07 | | Release date: | 2025-02-19 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Computational design of serine hydrolases.
Science, 388, 2025
|
|
6O0C
 
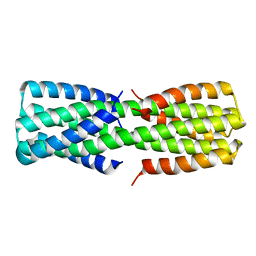 | | NMR ensemble of computationally designed protein XAA_GVDQ mutant M4L | | Descriptor: | Design construct XAA_GVDQ mutant M4L | | Authors: | Wei, K.Y, Moschidi, D, Nerli, S, Sgourakis, N, Baker, D. | | Deposit date: | 2019-02-15 | | Release date: | 2020-04-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Computational design of closely related proteins that adopt two well-defined but structurally divergent folds.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7LMX
 
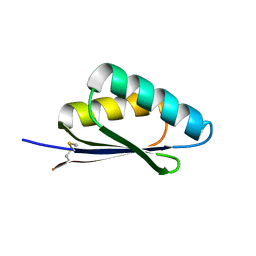 | | A HIGHLY SPECIFIC INHIBITOR OF INTEGRIN ALPHA-V BETA-6 WITH A DISULFIDE | | Descriptor: | Integrin inhibitor | | Authors: | Dong, X, Bera, A.K, Roy, A, Shi, L, Springer, T.A, Baker, D. | | Deposit date: | 2021-02-06 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | De novo design of highly selective miniprotein inhibitors of integrins alpha v beta 6 and alpha v beta 8.
Nat Commun, 14, 2023
|
|
7LMV
 
 | | SPECIFIC INHIBITOR OF INTEGRIN ALPHA-V BETA-6 | | Descriptor: | Integrin inhibitor | | Authors: | Dong, X, Bera, A.K, Roy, A, Shi, L, Springer, T.A, Baker, D. | | Deposit date: | 2021-02-05 | | Release date: | 2022-08-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | De novo design of highly selective miniprotein inhibitors of integrins alpha v beta 6 and alpha v beta 8.
Nat Commun, 14, 2023
|
|
3J9G
 
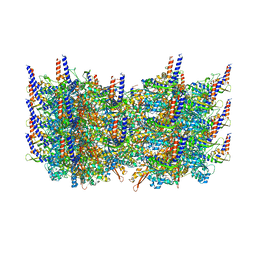 | | Atomic model of the VipA/VipB, the type six secretion system contractile sheath of Vibrio cholerae from cryo-EM | | Descriptor: | VipA, VipB | | Authors: | Kudryashev, M, Wang, R.Y.-R, Brackmann, M, Scherer, S, Maier, T, Baker, D, DiMaio, F, Stahlberg, H, Egelman, E.H, Basler, M. | | Deposit date: | 2015-01-16 | | Release date: | 2015-03-11 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the Type VI Secretion System Contractile Sheath.
Cell(Cambridge,Mass.), 160, 2015
|
|
3J1V
 
 | |
6DLM
 
 | | DHD127 | | Descriptor: | DHD127_A, DHD127_B | | Authors: | Bick, M.J, Chen, Z, Baker, D. | | Deposit date: | 2018-06-01 | | Release date: | 2018-12-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.753 Å) | | Cite: | Programmable design of orthogonal protein heterodimers.
Nature, 565, 2019
|
|
6DMA
 
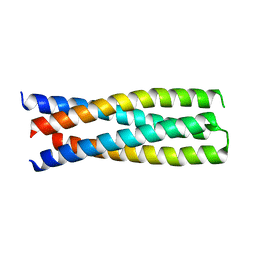 | | DHD15_closed | | Descriptor: | DHD15_closed_A, DHD15_closed_B | | Authors: | Bick, M.J, Chen, Z, Baker, D. | | Deposit date: | 2018-06-04 | | Release date: | 2018-12-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.363 Å) | | Cite: | Programmable design of orthogonal protein heterodimers.
Nature, 565, 2019
|
|
6DKM
 
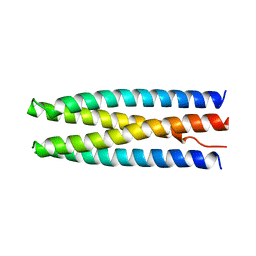 | | DHD131 | | Descriptor: | DHD131_A, DHD131_B | | Authors: | Bick, M.J, Chen, Z, Baker, D. | | Deposit date: | 2018-05-29 | | Release date: | 2018-12-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Programmable design of orthogonal protein heterodimers.
Nature, 565, 2019
|
|
6DM9
 
 | | DHD15_extended | | Descriptor: | DHD15_extended_A, DHD15_extended_B, SULFATE ION | | Authors: | Bick, M.J, Chen, Z, Baker, D. | | Deposit date: | 2018-06-04 | | Release date: | 2018-12-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Programmable design of orthogonal protein heterodimers.
Nature, 565, 2019
|
|
9NZH
 
 | |
5WP3
 
 | | Crystal Structure of EED in complex with EB22 | | Descriptor: | EB22, Polycomb protein EED, UNKNOWN ATOM OR ION | | Authors: | Dong, C, Tempel, W, Zhu, L, Moody, J.D, Baker, D, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-08-03 | | Release date: | 2017-09-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | First critical repressive H3K27me3 marks in embryonic stem cells identified using designed protein inhibitor.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4DRT
 
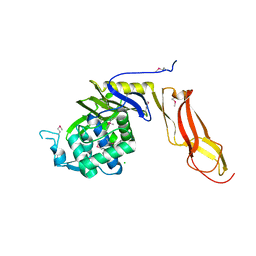 | | Three dimensional structure of de novo designed serine hydrolase OSH26, Northeast Structural Genomics Consortium (NESG) target OR89 | | Descriptor: | CHLORIDE ION, SODIUM ION, de novo designed serine hydrolase, ... | | Authors: | Kuzin, A, Su, M, Rajagopalan, S, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-02-17 | | Release date: | 2012-04-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
4D4E
 
 | | Crystal structure of computationally designed armadillo repeat proteins for modular peptide recognition. | | Descriptor: | ARMADILLO REPEAT PROTEIN ARM00016, GLYCEROL | | Authors: | Reichen, C, Forzani, C, Zhou, T, Parmeggiani, F, Fleishman, S.J, Mittl, P.R.E, Madhurantakam, C, Honegger, A, Ewald, C, Zerbe, O, Baker, D, Caflisch, A, Pluckthun, A. | | Deposit date: | 2014-10-28 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Computationally Designed Armadillo Repeat Proteins for Modular Peptide Recognition.
J.Mol.Biol., 428, 2016
|
|
4DDF
 
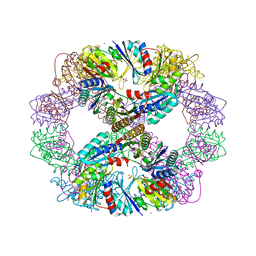 | | Computationally Designed Self-assembling Octahedral Cage protein, O333, Crystallized in space group P4 | | Descriptor: | CHLORIDE ION, Propanediol utilization polyhedral body protein PduT, SULFATE ION | | Authors: | Sawaya, M.R, King, N.P, Sheffler, W, Baker, D, Yeates, T.O. | | Deposit date: | 2012-01-18 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Computational design of self-assembling protein nanomaterials with atomic level accuracy.
Science, 336, 2012
|
|
6U1S
 
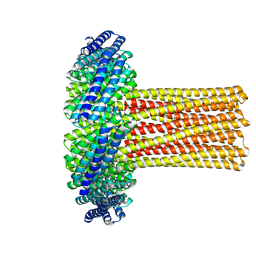 | | Cryo-EM structure of a de novo designed 16-helix transmembrane nanopore, TMHC8_R. | | Descriptor: | de novo designed 16-helix transmembrane nanopore, TMHC8_R | | Authors: | Johnson, M.J, Reggiano, G, Xu, C, Lu, P, Hsia, Y, Brunette, T.J, DiMaio, F, Baker, D, Kollman, J. | | Deposit date: | 2019-08-16 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Computational Design of Transmembrane Channels
To Be Published
|
|
4DCL
 
 | | Computationally Designed Self-assembling tetrahedron protein, T308, Crystallized in space group F23 | | Descriptor: | Putative acetyltransferase SACOL2570 | | Authors: | Sawaya, M.R, King, N.P, Sheffler, W, Baker, D, Yeates, T.O. | | Deposit date: | 2012-01-17 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Computational design of self-assembling protein nanomaterials with atomic level accuracy.
Science, 336, 2012
|
|
4LT9
 
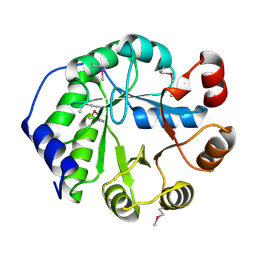 | | Crystal Structure of Engineered Protein, Northeast Structural Genomics Consortium Target OR404 | | Descriptor: | Engineered Protein OR404 | | Authors: | Vorobiev, S, Su, M, Bjelic, S, Kipnis, Y, Wang, L, Sahdev, S, Xiao, R, Kogan, S, Maglaqui, M, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-07-23 | | Release date: | 2013-08-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of Engineered Protein OR404.
To be Published
|
|
8VEI
 
 | |
8VEJ
 
 | | De novo designed cholic acid binder: CHD_buttress | | Descriptor: | CHD_buttress, CHOLIC ACID | | Authors: | Bera, A.K, Tran, L, Kang, A, Baker, D. | | Deposit date: | 2023-12-19 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Binding and sensing diverse small molecules using shape-complementary pseudocycles.
Science, 385, 2024
|
|
7CBC
 
 | |
8VEA
 
 | | De novo designed helical oligomer sg266 | | Descriptor: | Designed helical oligomer sg266 | | Authors: | Bera, A.K, Gerben, S, Baker, D. | | Deposit date: | 2023-12-18 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Massively parallel assessment of designed protein solution properties using mass spectrometry and peptide barcoding.
Biorxiv, 2025
|
|
