8D3Y
 
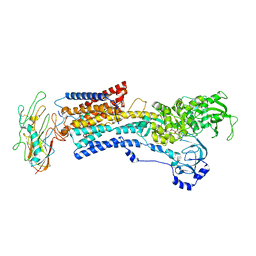 | | Human alpha3 Na+/K+-ATPase in its exoplasmic side-open state | | Descriptor: | FXYD domain-containing ion transport regulator 6, Sodium/potassium-transporting ATPase subunit alpha-3, Sodium/potassium-transporting ATPase subunit beta-1 | | Authors: | Nguyen, P.T, Bai, X. | | Deposit date: | 2022-06-01 | | Release date: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for gating mechanism of the human sodium-potassium pump.
Nat Commun, 13, 2022
|
|
8D3X
 
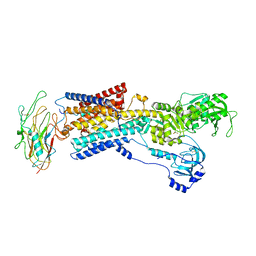 | | Human alpha3 Na+/K+-ATPase in its K+-occluded state | | Descriptor: | FXYD domain-containing ion transport regulator 6, Sodium/potassium-transporting ATPase subunit alpha-3, Sodium/potassium-transporting ATPase subunit beta-1, ... | | Authors: | Nguyen, P.T, Bai, X. | | Deposit date: | 2022-06-01 | | Release date: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis for gating mechanism of the human sodium-potassium pump.
Nat Commun, 13, 2022
|
|
8D3U
 
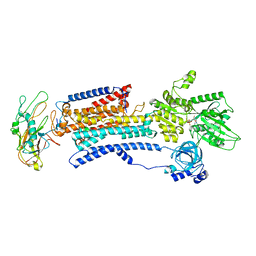 | | Human alpha3 Na+/K+-ATPase in its Na+-occluded state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FXYD domain-containing ion transport regulator 6, Sodium/potassium-transporting ATPase subunit alpha-3, ... | | Authors: | Nguyen, P.T, Bai, X. | | Deposit date: | 2022-06-01 | | Release date: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis for gating mechanism of the human sodium-potassium pump.
Nat Commun, 13, 2022
|
|
8D3W
 
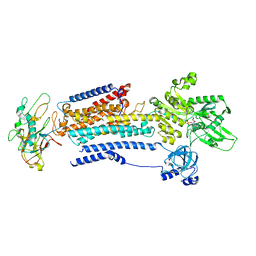 | |
8D3V
 
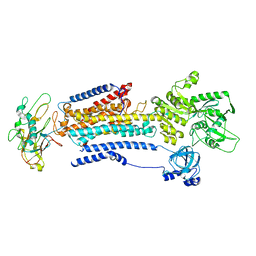 | | Human alpha3 Na+/K+-ATPase in its cytoplasmic side-open state | | Descriptor: | FXYD domain-containing ion transport regulator 6, Sodium/potassium-transporting ATPase subunit alpha-3, Sodium/potassium-transporting ATPase subunit beta-1 | | Authors: | Nguyen, P.T, Bai, X. | | Deposit date: | 2022-06-01 | | Release date: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for gating mechanism of the human sodium-potassium pump.
Nat Commun, 13, 2022
|
|
3J8H
 
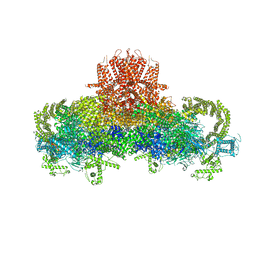 | | Structure of the rabbit ryanodine receptor RyR1 in complex with FKBP12 at 3.8 Angstrom resolution | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1A, Ryanodine receptor 1, ZINC ION | | Authors: | Yan, Z, Bai, X, Yan, C, Wu, J, Scheres, S.H.W, Shi, Y, Yan, N. | | Deposit date: | 2014-10-26 | | Release date: | 2014-12-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the rabbit ryanodine receptor RyR1 at near-atomic resolution.
Nature, 517, 2015
|
|
6U3U
 
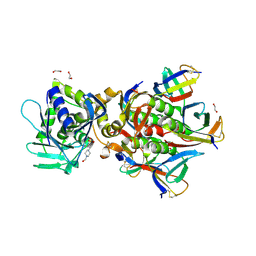 | | Crystal Structure of Shiga Toxin 2K | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Shiga toxin 2K subunit A, ... | | Authors: | Zhang, Y.Z, He, X.H. | | Deposit date: | 2019-08-22 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.287 Å) | | Cite: | Structural and Functional Characterization of Stx2k, a New Subtype of Shiga Toxin 2.
Microorganisms, 8, 2019
|
|
3BXW
 
 | |
7UDB
 
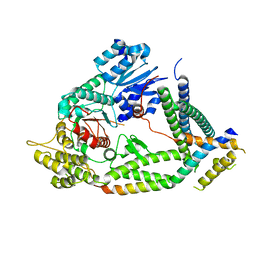 | | Cryo-EM structure of a synaptobrevin-Munc18-1-syntaxin-1 complex class 2 | | Descriptor: | Synaptobrevin-2, Syntaxin-1A, Syntaxin-binding protein 1 | | Authors: | Rizo, J, Bai, X, Stepien, K.P, Xu, J, Zhang, X. | | Deposit date: | 2022-03-18 | | Release date: | 2022-04-06 | | Last modified: | 2022-07-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | SNARE assembly enlightened by cryo-EM structures of a synaptobrevin-Munc18-1-syntaxin-1 complex.
Sci Adv, 8, 2022
|
|
7UDC
 
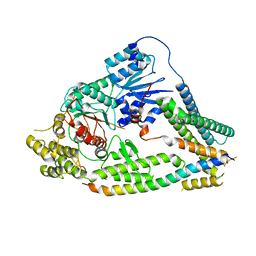 | | cryo-EM structures of a synaptobrevin-Munc18-1-syntaxin-1 complex class1 | | Descriptor: | Synaptobrevin-2, Syntaxin-1A, Syntaxin-binding protein 1 | | Authors: | Rizo, J, Bai, X, Stepien, K.P, Xu, J, Zhang, X. | | Deposit date: | 2022-03-18 | | Release date: | 2022-04-06 | | Last modified: | 2022-07-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | SNARE assembly enlightened by cryo-EM structures of a synaptobrevin-Munc18-1-syntaxin-1 complex.
Sci Adv, 8, 2022
|
|
7YTO
 
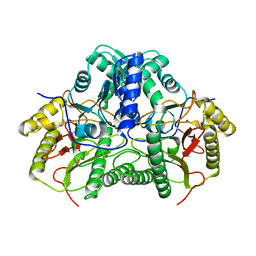 | |
1CP3
 
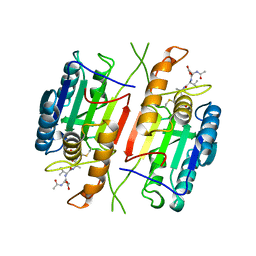 | |
6O84
 
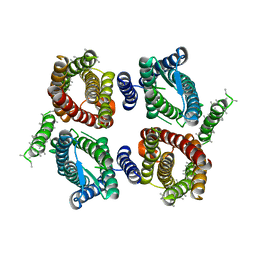 | | Cryo-EM structure of OTOP3 from xenopus tropicalis | | Descriptor: | LOC100127796 protein,LOC100127796 protein,OTOP3,LOC100127796 protein,LOC100127796 protein | | Authors: | Chen, Q.F, Bai, X.C, Jiang, Y.X. | | Deposit date: | 2019-03-08 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Structural and functional characterization of an otopetrin family proton channel.
Elife, 8, 2019
|
|
8EQB
 
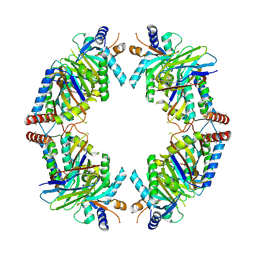 | | FAM46C/BCCIPalpha/Nanobody complex | | Descriptor: | Isoform 2 of BRCA2 and CDKN1A-interacting protein, Synthetic nanobody 1, Terminal nucleotidyltransferase 5C | | Authors: | Liu, S, Chen, H, Yin, Y, Bai, X, Zhang, X. | | Deposit date: | 2022-10-07 | | Release date: | 2023-03-15 | | Last modified: | 2023-04-26 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Inhibition of FAM46/TENT5 activity by BCCIP alpha adopting a unique fold.
Sci Adv, 9, 2023
|
|
2YBY
 
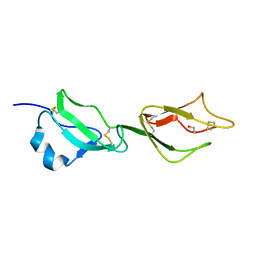 | | Structure of domains 6 and 7 of the mouse complement regulator Factor H | | Descriptor: | 1,2-ETHANEDIOL, COMPLEMENT FACTOR H | | Authors: | Everett, R.J, Caesar, J.J.E, Johnson, S.J, Tang, C.M, Lea, S.M. | | Deposit date: | 2011-03-30 | | Release date: | 2012-04-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Design and Evaluation of Meningococcal Vaccines Through Structure-Based Modification of Host and Pathogen Molecules.
Plos Pathog., 8, 2012
|
|
5JSJ
 
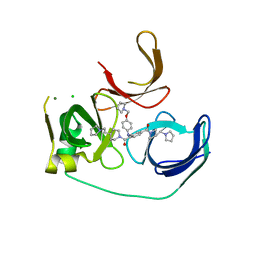 | | Crystal structure of Spindlin1 bound to compound EML631 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Spindlin-1, ... | | Authors: | Su, X, Li, H. | | Deposit date: | 2016-05-08 | | Release date: | 2017-05-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.348 Å) | | Cite: | Developing Spindlin1 small-molecule inhibitors by using protein microarrays
Nat. Chem. Biol., 13, 2017
|
|
6C9A
 
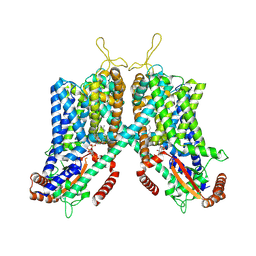 | | Cryo-EM structure of mouse TPC1 channel in the PtdIns(3,5)P2-bound state | | Descriptor: | (2R)-3-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,4,6-trihydroxy-3,5-bis(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dioctanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | She, J, Guo, J, Chen, Q, Bai, X, Jiang, Y. | | Deposit date: | 2018-01-25 | | Release date: | 2018-04-04 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into the voltage and phospholipid activation of the mammalian TPC1 channel.
Nature, 556, 2018
|
|
6C96
 
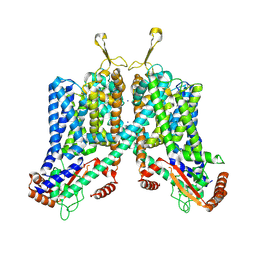 | | Cryo-EM structure of mouse TPC1 channel in the apo state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SODIUM ION, ... | | Authors: | She, J, Guo, J, Chen, Q, Bai, X, Jiang, Y. | | Deposit date: | 2018-01-25 | | Release date: | 2018-04-04 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into the voltage and phospholipid activation of the mammalian TPC1 channel.
Nature, 556, 2018
|
|
5JSG
 
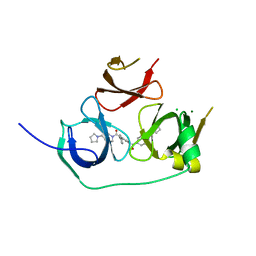 | | Crystal structure of Spindlin1 bound to compound EML405 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Spindlin-1, ... | | Authors: | Su, X, Li, H. | | Deposit date: | 2016-05-08 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Developing Spindlin1 small-molecule inhibitors by using protein microarrays
Nat. Chem. Biol., 13, 2017
|
|
6O58
 
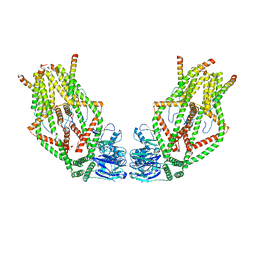 | | Human MCU-EMRE complex, dimer of channel | | Descriptor: | CALCIUM ION, Calcium uniporter protein, mitochondrial, ... | | Authors: | Wang, Y, Bai, X, Jiang, Y. | | Deposit date: | 2019-03-01 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural Mechanism of EMRE-Dependent Gating of the Human Mitochondrial Calcium Uniporter.
Cell, 177, 2019
|
|
6O5B
 
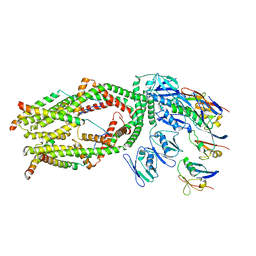 | | Monomer of a cation channel | | Descriptor: | CALCIUM ION, Calcium uniporter protein, mitochondrial, ... | | Authors: | Wang, Y, Bai, X, Jiang, Y. | | Deposit date: | 2019-03-01 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural Mechanism of EMRE-Dependent Gating of the Human Mitochondrial Calcium Uniporter.
Cell, 177, 2019
|
|
3JBT
 
 | | Atomic structure of the Apaf-1 apoptosome | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Apoptotic protease-activating factor 1, Cytochrome c, ... | | Authors: | Zhou, M, Li, Y, Hu, Q, Bai, X, Huang, W, Yan, C, Scheres, S.H.W, Shi, Y. | | Deposit date: | 2015-10-15 | | Release date: | 2015-11-18 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Atomic structure of the apoptosome: mechanism of cytochrome c- and dATP-mediated activation of Apaf-1
Genes Dev., 29, 2015
|
|
3J9K
 
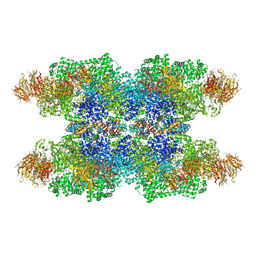 | | Structure of Dark apoptosome in complex with Dronc CARD domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Apaf-1 related killer DARK, Caspase Nc | | Authors: | Pang, Y, Bai, X, Yan, C, Hao, Q, Chen, Z, Wang, J, Scheres, S.H.W, Shi, Y. | | Deposit date: | 2015-02-04 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of the apoptosome: mechanistic insights into activation of an initiator caspase from Drosophila.
Genes Dev., 29, 2015
|
|
3J9L
 
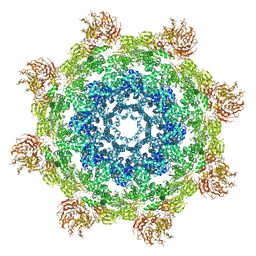 | | Structure of Dark apoptosome from Drosophila melanogaster | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Apaf-1 related killer DARK | | Authors: | Pang, Y, Bai, X, Yan, C, Hao, Q, Chen, Z, Wang, J, Scheres, S.H.W, Shi, Y. | | Deposit date: | 2015-02-04 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of the apoptosome: mechanistic insights into activation of an initiator caspase from Drosophila.
Genes Dev., 29, 2015
|
|
5WPT
 
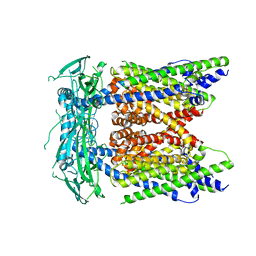 | | Cryo-EM structure of mammalian endolysosomal TRPML1 channel in nanodiscs in closed II conformation at 3.75 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Mucolipin-1, SODIUM ION | | Authors: | Chen, Q, She, J, Guo, J, Bai, X, Jiang, Y. | | Deposit date: | 2017-08-07 | | Release date: | 2017-10-18 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Structure of mammalian endolysosomal TRPML1 channel in nanodiscs.
Nature, 550, 2017
|
|
