4LCD
 
 | |
5DG5
 
 | | CRYSTAL STRUCTURE OF THE TYROSINE KINASE DOMAIN OF THE HEPATOCYTE GROWTH FACTOR RECEPTOR C-MET IN COMPLEX WITH ALTIRATINIB ANALOG DP-4157 | | Descriptor: | Hepatocyte growth factor receptor, N-(2,5-difluoro-4-{[2-(1-methyl-1H-pyrazol-4-yl)pyridin-4-yl]oxy}phenyl)-N'-(4-fluorophenyl)cyclopropane-1,1-dicarboxam ide | | Authors: | Smith, B.D, Kaufman, M.D, Leary, C.B, Turner, B.A, Wise, S.A, Ahn, Y.M, Booth, R.J, Caldwell, T.M, Ensinger, C.L, Hood, M.M, Lu, W.-P, Patt, T.W, Patt, W.C, Rutkoski, T.J, Samarakoon, T, Telikepalli, H, Vogeti, L, Vogeti, S, Yates, K.M, Chun, L, Stewart, L.J, Clare, M, Flynn, D.L. | | Deposit date: | 2015-08-27 | | Release date: | 2016-08-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Altiratinib Inhibits Tumor Growth, Invasion, Angiogenesis, and Microenvironment-Mediated Drug Resistance via Balanced Inhibition of MET, TIE2, and VEGFR2.
Mol.Cancer Ther., 14, 2015
|
|
4LND
 
 | | Crystal structure of human apurinic/apyrimidinic endonuclease 1 with essential Mg2+ cofactor | | Descriptor: | DNA-(apurinic or apyrimidinic site) lyase, MAGNESIUM ION | | Authors: | Manvilla, B.A, Pozharski, E, Toth, E.A, Drohat, A.C. | | Deposit date: | 2013-07-11 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure of human apurinic/apyrimidinic endonuclease 1 with the essential Mg(2+) cofactor.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5DVA
 
 | |
5DN4
 
 | |
5DTT
 
 | | Fragments bound to the OXA-48 beta-lactamase: Compound 3 | | Descriptor: | 1,2-ETHANEDIOL, 3-(1,3-thiazol-2-yl)benzoic acid, Beta-lactamase, ... | | Authors: | Lund, B.A, Christopeit, T, Leiros, H.-K.S. | | Deposit date: | 2015-09-18 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.10000539 Å) | | Cite: | Screening and Design of Inhibitor Scaffolds for the Antibiotic Resistance Oxacillinase-48 (OXA-48) through Surface Plasmon Resonance Screening.
J.Med.Chem., 59, 2016
|
|
5DTS
 
 | |
5E0U
 
 | |
4LS9
 
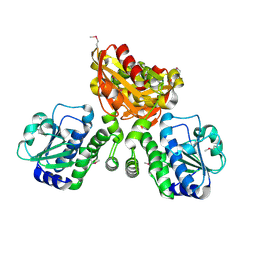 | | Structure of mycobacterial nrnA homolog reveals multifunctional nuclease activities | | Descriptor: | DHH family protein, GLYCEROL, MANGANESE (II) ION | | Authors: | Kumar, D, Srivastav, R, Grover, A, Manjasetty, B.A, Sharma, R, Taneja, B. | | Deposit date: | 2013-07-22 | | Release date: | 2014-07-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Unique subunit packing in mycobacterial nanoRNase leads to alternate substrate recognitions in DHH phosphodiesterases
Nucleic Acids Res., 42, 2014
|
|
4M18
 
 | | Crystal Structure of Surfactant Protein-D D325A/R343V mutant in complex with trimannose (Man-a1,2Man-a1,2Man) | | Descriptor: | CALCIUM ION, Pulmonary surfactant-associated protein D, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose, ... | | Authors: | Goh, B.C, Rynkiewicz, M.J, Cafarella, T.R, White, M.R, Hartshorn, K.L, Allen, K, Crouch, E.C, Calin, O, Seeberger, P.H, Schulten, K, Seaton, B.A. | | Deposit date: | 2013-08-02 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.203 Å) | | Cite: | Molecular mechanisms of inhibition of influenza by surfactant protein d revealed by large-scale molecular dynamics simulation.
Biochemistry, 52, 2013
|
|
5E0V
 
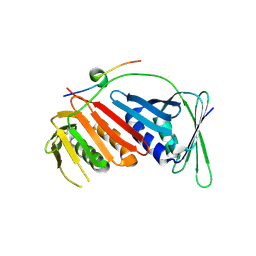 | |
5E7N
 
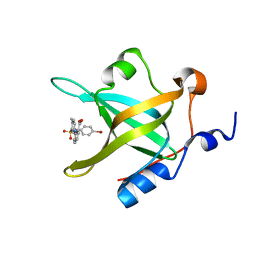 | | Crystal Structure of RPA70N in complex with VU0085636 | | Descriptor: | 2-({3-[(4-bromophenyl)sulfamoyl]-4-methylbenzoyl}amino)benzoic acid, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Gilston, B.A, Patrone, J.D, Pelz, N.F, Bates, B.S, Souza-Fagundes, E.M, Vangamudi, B, Camper, D, Kuznetsov, A, Browning, C.F, Feldkamp, M.D, Olejniczak, E.T, Rossanese, O.W, Waterson, A.G, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2015-10-12 | | Release date: | 2016-01-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Identification and Optimization of Anthranilic Acid Based Inhibitors of Replication Protein A.
Chemmedchem, 11, 2016
|
|
5DN5
 
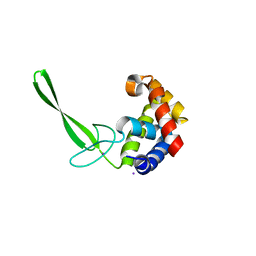 | | Structure of a C-terminally truncated glycoside hydrolase domain from Salmonella typhimurium FlgJ | | Descriptor: | CHLORIDE ION, IODIDE ION, Peptidoglycan hydrolase FlgJ, ... | | Authors: | Zaloba, P, Bailey-Elkin, B.A, Mark, B.L. | | Deposit date: | 2015-09-09 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and Biochemical Insights into the Peptidoglycan Hydrolase Domain of FlgJ from Salmonella typhimurium.
Plos One, 11, 2016
|
|
4LP0
 
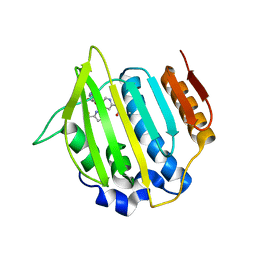 | | Crystal structure of a topoisomerase ATP inhibitor | | Descriptor: | 6'-[(ethylcarbamoyl)amino]-4'-[4-(trifluoromethyl)-1,3-thiazol-2-yl]-3,3'-bipyridine-5-carboxylic acid, Topoisomerase IV subunit B | | Authors: | Basarab, G.S, Manchester, J.I, Bist, S, Boriack-Sjodin, P.A, Dangel, B, Illingsworth, R, Uria-Nickelsen, M, Sherer, B.A, Sriram, S, Eakin, A.E. | | Deposit date: | 2013-07-14 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Fragment-to-Hit-to-Lead Discovery of a Novel Pyridylurea Scaffold of ATP Competitive Dual Targeting Type II Topoisomerase Inhibiting Antibacterial Agents.
J.Med.Chem., 56, 2013
|
|
5E0T
 
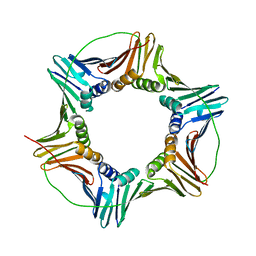 | | Human PCNA mutant - S228I | | Descriptor: | Proliferating cell nuclear antigen | | Authors: | Duffy, C.M, Hilbert, B.J, Kelch, B.A. | | Deposit date: | 2015-09-29 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6653 Å) | | Cite: | A Disease-Causing Variant in PCNA Disrupts a Promiscuous Protein Binding Site.
J.Mol.Biol., 428, 2016
|
|
4M17
 
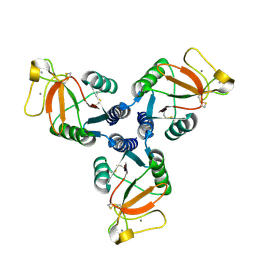 | | Crystal Structure of Surfactant Protein-D D325A/R343V mutant | | Descriptor: | CALCIUM ION, Pulmonary surfactant-associated protein D | | Authors: | Goh, B.C, Rynkiewicz, M.J, Cafarella, T.R, White, M.R, Hartshorn, K.L, Allen, K, Crouch, E.C, Calin, O, Seeberger, P.H, Schulten, K, Seaton, B.A. | | Deposit date: | 2013-08-02 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.096 Å) | | Cite: | Molecular mechanisms of inhibition of influenza by surfactant protein d revealed by large-scale molecular dynamics simulation.
Biochemistry, 52, 2013
|
|
5DTK
 
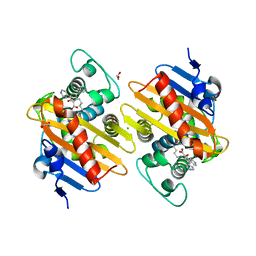 | | Fragments bound to the OXA-48 beta-lactamase: Compound 17 | | Descriptor: | 1,2-ETHANEDIOL, 3,5-di(pyridin-4-yl)benzoic acid, Beta-lactamase, ... | | Authors: | Lund, B.A, Christopeit, T, Leiros, H.-K.S. | | Deposit date: | 2015-09-18 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.60000241 Å) | | Cite: | Screening and Design of Inhibitor Scaffolds for the Antibiotic Resistance Oxacillinase-48 (OXA-48) through Surface Plasmon Resonance Screening.
J.Med.Chem., 59, 2016
|
|
5N13
 
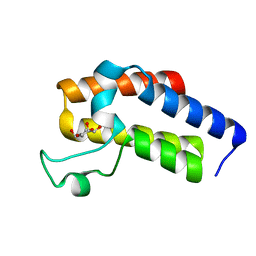 | | Second Bromodomain (BD2) from Candida albicans Bdf1 in the unbound form | | Descriptor: | Bromodomain-containing factor 1, GLYCEROL | | Authors: | Mietton, F, Ferri, E, Champlebouxm, M, Zala, N, Maubon, D, Zhou, Y, Harbut, M, Spittler, D, Garnaud, C, Courcon, M, Chauvel, M, d'Enfert, C, Kashemirov, B.A, Hull, M, Cornet, M, McKenna, C.E, Govin, J, Petosa, C. | | Deposit date: | 2017-02-04 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Selective BET bromodomain inhibition as an antifungal therapeutic strategy.
Nat Commun, 8, 2017
|
|
5N17
 
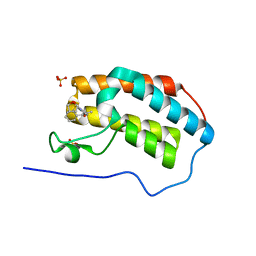 | | First Bromodomain (BD1) from Candida albicans Bdf1 bound to a dibenzothiazepinone (compound 3) | | Descriptor: | (2~{S})-~{N}-(5-methyl-6-oxidanylidene-benzo[b][1,4]benzothiazepin-2-yl)oxolane-2-carboxamide, Bromodomain-containing factor 1, SULFATE ION | | Authors: | Mietton, F, Ferri, E, Champleboux, M, Zala, N, Maubon, D, Zhou, Y, Harbut, M, Spittler, D, Garnaud, C, Courcon, M, Chauvon, M, d'Enfer, C, Kashemirov, B.A, Hull, M, Cornet, M, McKenna, C.E, Govin, J, Petosa, C. | | Deposit date: | 2017-02-05 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Selective BET bromodomain inhibition as an antifungal therapeutic strategy.
Nat Commun, 8, 2017
|
|
5N15
 
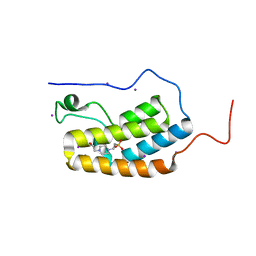 | | First Bromodomain (BD1) from Candida albicans Bdf1 in the unbound form | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Bromodomain-containing factor 1, GLYCEROL, ... | | Authors: | Mietton, F, Ferri, E, Champleboux, M, Zala, N, Maubon, D, Zhou, Y, Harbut, M, Spittler, D, Garnaud, C, Courcon, M, Chauvel, M, d'Enfert, C, Kashemirov, B.A, Hull, M, Cornet, M, McKenna, C.E, Govin, J, Petosa, C. | | Deposit date: | 2017-02-05 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Selective BET bromodomain inhibition as an antifungal therapeutic strategy.
Nat Commun, 8, 2017
|
|
5N18
 
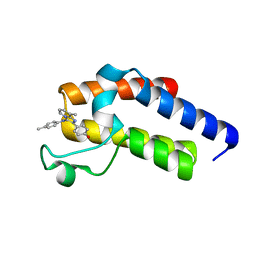 | | Second Bromodomain (BD2) from Candida albicans Bdf1 bound to an imidazopyridine (compound 2) | | Descriptor: | 4-[8-methyl-3-[(4-methylphenyl)amino]imidazo[1,2-a]pyridin-2-yl]phenol, Bromodomain-containing factor 1, GLYCEROL | | Authors: | Mietton, F, Ferri, E, Champleboux, M, Zala, N, Maubon, D, Zhou, Y, Harbut, M, Spittler, D, Garnaud, C, Courcon, M, Chauvel, M, d'Enfert, C, Kashemirov, B.A, Hull, M, Cornet, M, McKenna, C.E, Govin, J, Petosa, C. | | Deposit date: | 2017-02-05 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Selective BET bromodomain inhibition as an antifungal therapeutic strategy.
Nat Commun, 8, 2017
|
|
5OE2
 
 | | CRYSTAL STRUCTURE OF THE BETA-LACTAMASE OXA-245 | | Descriptor: | Beta-lactamase, CHLORIDE ION, SODIUM ION | | Authors: | Lund, B.A, Leiros, H.K.S. | | Deposit date: | 2017-07-07 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure, activity and thermostability investigations of OXA-163, OXA-181 and OXA-245 using biochemical analysis, crystal structures and differential scanning calorimetry analysis.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
5OE0
 
 | | CRYSTAL STRUCTURE OF THE BETA-LACTAMASE OXA-181 | | Descriptor: | Beta-lactamase, CHLORIDE ION, SULFATE ION | | Authors: | Lund, B.A, Carlsen, T.J.O, Leiros, H.K.S, Thomassen, A.M. | | Deposit date: | 2017-07-07 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.0500083 Å) | | Cite: | Structure, activity and thermostability investigations of OXA-163, OXA-181 and OXA-245 using biochemical analysis, crystal structures and differential scanning calorimetry analysis.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
5ODZ
 
 | | CRYSTAL STRUCTURE OF THE BETA-LACTAMASE OXA-163 | | Descriptor: | Beta-lactamase, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Lund, B.A, Carlsen, T.J.O, Leiros, H.K.S. | | Deposit date: | 2017-07-07 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure, activity and thermostability investigations of OXA-163, OXA-181 and OXA-245 using biochemical analysis, crystal structures and differential scanning calorimetry analysis.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
5OFT
 
 | |
