6SG4
 
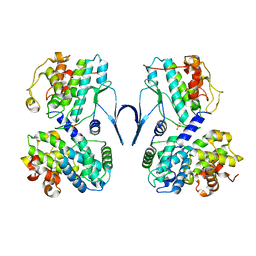 | | Structure of CDK2/cyclin A M246Q, S247EN | | Descriptor: | Cyclin-A2, Cyclin-dependent kinase 2 | | Authors: | Salamina, M, Basle, A, Massa, B, Noble, M.E.M, Endicott, J.A. | | Deposit date: | 2019-08-02 | | Release date: | 2021-01-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Discriminative SKP2 Interactions with CDK-Cyclin Complexes Support a Cyclin A-Specific Role in p27KIP1 Degradation.
J.Mol.Biol., 433, 2021
|
|
4LS3
 
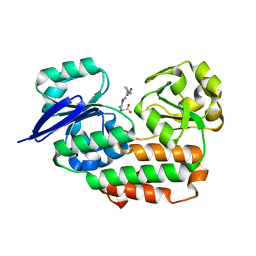 | | THE crystal STRUCTURE OF HELICOBACTER PYLORI CEUE(HP1561)/NI-HIS COMPL | | Descriptor: | HISTIDINE, NICKEL (II) ION, Nickel (III) ABC transporter, ... | | Authors: | Salamina, M, Shaik, M.M, Cendron, L, Zanotti, G. | | Deposit date: | 2013-07-22 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Helicobacter pylori periplasmic receptor CeuE (HP1561) modulates its nickel affinity via organic metallophores.
Mol.Microbiol., 91, 2014
|
|
2VEE
 
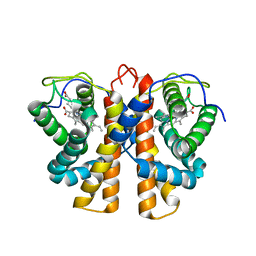 | | Structure of protoglobin from Methanosarcina acetivorans C2A | | Descriptor: | PROTOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nardini, M, Pesce, A, Thijs, L, Saito, J.A, Dewilde, S, Alam, M, Ascenzi, P, Coletta, M, Ciaccio, C, Moens, L, Bolognesi, M. | | Deposit date: | 2007-10-22 | | Release date: | 2008-01-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Archaeal Protoglobin Structure Indicates New Ligand Diffusion Paths and Modulation of Haem-Reactivity.
Embo Rep., 9, 2008
|
|
2VEB
 
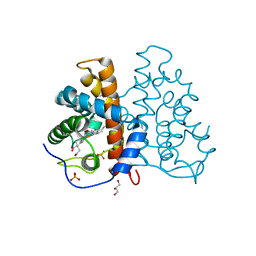 | | High resolution structure of protoglobin from Methanosarcina acetivorans C2A | | Descriptor: | GLYCEROL, OXYGEN MOLECULE, PHOSPHATE ION, ... | | Authors: | Nardini, M, Pesce, A, Thijs, L, Saito, J.A, Dewilde, S, Alam, M, Ascenzi, P, Coletta, M, Ciaccio, C, Moens, L, Bolognesi, M. | | Deposit date: | 2007-10-18 | | Release date: | 2008-01-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Archaeal Protoglobin Structure Indicates New Ligand Diffusion Paths and Modulation of Haem-Reactivity.
Embo Rep., 9, 2008
|
|
2W31
 
 | | globin domain of Geobacter sulfurreducens globin-coupled sensor | | Descriptor: | GLOBIN, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Pesce, A, Thijs, L, Nardini, M, Desmet, F, Sisinni, L, Gourlay, L, Bolli, A, Coletta, M, Van Doorslaer, S, Wan, X, Alam, M, Ascenzi, P, Moens, L, Bolognesi, M, Dewilde, S. | | Deposit date: | 2008-11-05 | | Release date: | 2009-01-13 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Hise11 and Hisf8 Provide Bis-Histidyl Heme Hexa-Coordination in the Globin Domain of Geobacter Sulfurreducens Globin-Coupled Sensor.
J.Mol.Biol., 386, 2009
|
|
4E2O
 
 | | Crystal structure of alpha-amylase from Geobacillus thermoleovorans, GTA, complexed with acarbose | | Descriptor: | 6-AMINO-4-HYDROXYMETHYL-CYCLOHEX-4-ENE-1,2,3-TRIOL, Alpha-amylase, CALCIUM ION, ... | | Authors: | Mok, S.C, Teh, A.H, Saito, J.A, Najimudin, N, Alam, M. | | Deposit date: | 2012-03-09 | | Release date: | 2013-03-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Crystal structure of a compact alpha-amylase from Geobacillus thermoleovorans.
Enzyme.Microb.Technol., 53, 2013
|
|
3S1J
 
 | | Crystal structure of acetate-bound hell's gate globin I | | Descriptor: | ACETATE ION, Hemoglobin-like flavoprotein, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Teh, A.H, Saito, J.A, Baharuddin, A, Tuckerman, J.R, Newhouse, J.S, Kanbe, M, Newhouse, E.I, Rahim, R.A, Favier, F, Didierjean, C, Sousa, E.H.S, Stott, M.B, Dunfield, P.F, Gonzalez, G, Gilles-Gonzalez, M.A, Najimudin, N, Alam, M. | | Deposit date: | 2011-05-15 | | Release date: | 2011-09-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Hell's Gate globin I: an acid and thermostable bacterial hemoglobin resembling mammalian neuroglobin
Febs Lett., 585, 2011
|
|
3S1I
 
 | | Crystal structure of oxygen-bound hell's gate globin I | | Descriptor: | Hemoglobin-like flavoprotein, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Teh, A.H, Saito, J.A, Baharuddin, A, Tuckerman, J.R, Newhouse, J.S, Kanbe, M, Newhouse, E.I, Rahim, R.A, Favier, F, Didierjean, C, Sousa, E.H.S, Stott, M.B, Dunfield, P.F, Gonzalez, G, Gilles-Gonzalez, M.A, Najimudin, N, Alam, M. | | Deposit date: | 2011-05-15 | | Release date: | 2011-09-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Hell's Gate globin I: an acid and thermostable bacterial hemoglobin resembling mammalian neuroglobin
Febs Lett., 585, 2011
|
|
7PKY
 
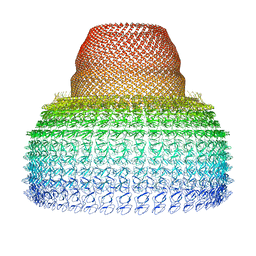 | | Half-vault structure | | Descriptor: | Major vault protein | | Authors: | Guerra, P, Gonzalez-Alamos, M, Llauro, A, Casanas, A, Querol-Audi, J, de Pablo, P, Verdaguer, N. | | Deposit date: | 2021-08-27 | | Release date: | 2022-03-23 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Symmetry disruption commits vault particles to disassembly.
Sci Adv, 8, 2022
|
|
7PKR
 
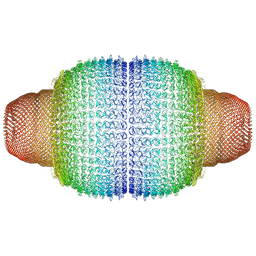 | | Vault structure in primmed conformation | | Descriptor: | Major vault protein | | Authors: | Guerra, P, Gonzalez-Alamos, M, Llauro, A, Casanas, A, Querol-Audi, J, de Pablo, P, Verdaguer, N. | | Deposit date: | 2021-08-26 | | Release date: | 2022-03-16 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Symmetry disruption commits vault particles to disassembly.
Sci Adv, 8, 2022
|
|
7PKZ
 
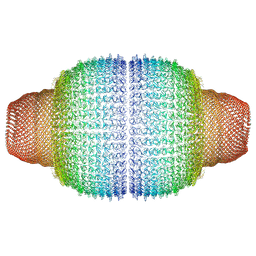 | | Vault structure in committed conformation | | Descriptor: | Major vault protein | | Authors: | Guerra, P, Gonzalez-Alamos, M, Llauro, A, Casanas, A, Querol-Audi, J, de Pablo, P, Verdaguer, N. | | Deposit date: | 2021-08-27 | | Release date: | 2022-03-16 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (9.8 Å) | | Cite: | Symmetry disruption commits vault particles to disassembly.
Sci Adv, 8, 2022
|
|
7TNG
 
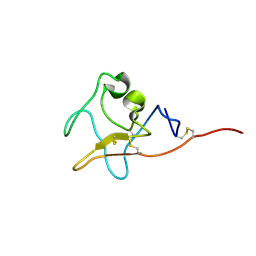 | | Kringle domain of human Receptor Tyrosine Kinase-Like Orphan Receptor 1 (ROR1) | | Descriptor: | Inactive tyrosine-protein kinase transmembrane receptor ROR1 | | Authors: | Guarino, S.R, Di Bello, A, Palamini, M, Forneris, F. | | Deposit date: | 2022-01-21 | | Release date: | 2022-05-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the kringle domain of human receptor tyrosine kinase-like orphan receptor 1 (hROR1)
Acta Crystallogr.,Sect.F, 78, 2022
|
|
8BZO
 
 | | Cryo-EM structure of CDK2-CyclinA in complex with p27 from the SCFSKP2 E3 ligase Complex | | Descriptor: | Cyclin-A2, Cyclin-dependent kinase 2, Cyclin-dependent kinase inhibitor 1B | | Authors: | Rowland, R.J, Salamina, M, Endicott, J.A, Noble, M.E. | | Deposit date: | 2022-12-15 | | Release date: | 2023-06-28 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of SKP1-SKP2-CKS1 in complex with CDK2-cyclin A-p27KIP1.
Sci Rep, 13, 2023
|
|
8BYA
 
 | | Cryo-EM structure of SKP1-SKP2-CKS1-CDK2-CyclinA-p27KIP1 Complex | | Descriptor: | Cyclin-A2, Cyclin-dependent kinase 2, Cyclin-dependent kinase inhibitor 1B, ... | | Authors: | Rowland, R.J, Salamina, M, Endicott, J.A, Noble, M.E. | | Deposit date: | 2022-12-12 | | Release date: | 2023-06-28 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Cryo-EM structure of SKP1-SKP2-CKS1 in complex with CDK2-cyclin A-p27KIP1.
Sci Rep, 13, 2023
|
|
8BYL
 
 | | Cryo-EM structure of SKP1-SKP2-CKS1 from the SCFSKP2 E3 ligase complex | | Descriptor: | Cyclin-dependent kinase inhibitor 1B, Cyclin-dependent kinases regulatory subunit 1, S-phase kinase-associated protein 1, ... | | Authors: | Rowland, R.J, Salamina, M, Endicott, J.A, Noble, M.E.M. | | Deposit date: | 2022-12-13 | | Release date: | 2023-06-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of SKP1-SKP2-CKS1 in complex with CDK2-cyclin A-p27KIP1.
Sci Rep, 13, 2023
|
|
1JS1
 
 | | Crystal Structure of a new transcarbamylase from the anaerobic bacterium Bacteroides fragilis at 2.0 A resolution | | Descriptor: | PHOSPHATE ION, Transcarbamylase | | Authors: | Shi, D, Gallegos, R, DePonte III, J, Morizono, H, Yu, X, Allewell, N.M, Malamy, M, Tuchman, M. | | Deposit date: | 2001-08-15 | | Release date: | 2002-07-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a transcarbamylase-like protein from the anaerobic bacterium Bacteroides fragilis at 2.0 A resolution.
J.Mol.Biol., 320, 2002
|
|
2JO0
 
 | | The solution structure of the monomeric species of the C terminal domain of the CA protein of HIV-1 | | Descriptor: | Gag-Pol polyprotein | | Authors: | Alcaraz, L.A, del Alamo, M, Barrera, F.N, Mateu, M.G, Neira, J.L. | | Deposit date: | 2007-02-17 | | Release date: | 2007-07-31 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Flexibility in HIV-1 Assembly Subunits: Solution Structure of the Monomeric C-Terminal Domain of the Capsid Protein
Biophys.J., 93, 2007
|
|
4ZZF
 
 | | Crystal structure of truncated FlgD (tetragonal form) from the human pathogen Helicobacter pylori | | Descriptor: | Flagellar basal body rod modification protein | | Authors: | Pulic, I, Cendron, L, Salamina, M, Matkovic-Calogovic, D, Zanotti, G. | | Deposit date: | 2015-05-22 | | Release date: | 2016-02-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1673 Å) | | Cite: | Crystal structure of truncated FlgD from the human pathogen Helicobacter pylori.
J.Struct.Biol., 194, 2016
|
|
4ZZK
 
 | | Crystal structure of truncated FlgD (monoclinic form) from the human pathogen Helicobacter pylori | | Descriptor: | Basal-body rod modification protein FlgD | | Authors: | Pulic, I, Cendron, L, Salamina, M, Matkovic-Calogovic, D, Zanotti, G. | | Deposit date: | 2015-05-22 | | Release date: | 2016-02-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of truncated FlgD from the human pathogen Helicobacter pylori.
J.Struct.Biol., 194, 2016
|
|
2P33
 
 | | Synthesis and SAR of Aminopyrimidines as Novel c-Jun N-Terminal Kinase (JNK) Inhibitors | | Descriptor: | 4-{[5-chloro-4-(1H-indol-3-yl)pyrimidin-2-yl]amino}-N-ethylpiperidine-1-carboxamide, c-Jun N-terminal kinase 3 | | Authors: | Ceska, T.A, Platt, A, Fortunato, M, Dickson, K.M, Sharpe, A. | | Deposit date: | 2007-03-08 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Synthesis and SAR of aminopyrimidines as novel c-Jun N-terminal kinase (JNK) inhibitors
Bioorg.Med.Chem.Lett., 17, 2007
|
|
3WFX
 
 | | Crystal Structure of the Imidazole-Bound Form of the HGbRL's Globin Domain | | Descriptor: | HEXANE-1,6-DIOL, Hemoglobin-like flavoprotein fused to Roadblock/LC7 domain, IMIDAZOLE, ... | | Authors: | Teh, A.H. | | Deposit date: | 2013-07-24 | | Release date: | 2014-08-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Open and Lys-His Hexacoordinated Closed Structures of a Globin with Swapped Proximal and Distal Sites.
Sci Rep, 5, 2015
|
|
6GVA
 
 | | CDK2/cyclin A2 in complex with pyrazolo[4,3-d]pyrimidine inhibitor LGR4455 | | Descriptor: | 1,2-ETHANEDIOL, 5-(2-azanylethylsulfanyl)-3-propan-2-yl-~{N}-[(4-pyridin-2-ylphenyl)methyl]-2~{H}-pyrazolo[4,3-d]pyrimidin-7-amine, BROMIDE ION, ... | | Authors: | Skerlova, J, Rezacova, P. | | Deposit date: | 2018-06-20 | | Release date: | 2019-05-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | 3,5,7-Substituted Pyrazolo[4,3- d]pyrimidine Inhibitors of Cyclin-Dependent Kinases and Their Evaluation in Lymphoma Models.
J.Med.Chem., 62, 2019
|
|
2VW5
 
 | | Structure Of The Hsp90 Inhibitor 7-O-carbamoylpremacbecin Bound To The N- Terminus Of Yeast Hsp90 | | Descriptor: | (4E,8S,9R,10E,12S,13R,14S,16R)-13,20-dihydroxy-14-methoxy-4,8,10,12,16-pentamethyl-3-oxo-2-azabicyclo[16.3.1]docosa-1(22),4,10,18,20-pentaen-9-yl carbamate, ATP-DEPENDENT MOLECULAR CHAPERONE HSP82, SULFATE ION | | Authors: | Roe, S.M, Prodromou, C, Pearl, L.H. | | Deposit date: | 2008-06-16 | | Release date: | 2008-09-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Optimizing Natural Products by Biosynthetic Engineering: Discovery of Nonquinone Hsp90 Inhibitors.
J.Med.Chem., 51, 2008
|
|
4TOY
 
 | |
4NK2
 
 | | Crystal structure of Hell's gate globin IV | | Descriptor: | Hemoglobin-like protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Jamil, F. | | Deposit date: | 2013-11-12 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure of truncated haemoglobin from an extremely thermophilic and acidophilic bacterium.
J.Biochem., 156, 2014
|
|
