2MNG
 
 | | Apo Structure of human HCN4 CNBD solved by NMR | | 分子名称: | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 | | 著者 | Akimoto, M, Zhang, Z, Boulton, S, Selvaratnam, R, VanSchouwen, B, Gloyd, M, Accili, E.A, Lange, O.F, Melacini, G. | | 登録日 | 2014-04-03 | | 公開日 | 2014-06-04 | | 最終更新日 | 2015-02-11 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A mechanism for the auto-inhibition of hyperpolarization-activated cyclic nucleotide-gated (HCN) channel opening and its relief by cAMP.
J.Biol.Chem., 289, 2014
|
|
7D5V
 
 | | Structure of the C646A mutant of peptidylarginine deiminase type III (PAD3) | | 分子名称: | 1,2-ETHANEDIOL, GLYCEROL, Protein-arginine deiminase type-3 | | 著者 | Akimoto, M, Mashimo, R, Unno, M. | | 登録日 | 2020-09-28 | | 公開日 | 2021-06-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.102 Å) | | 主引用文献 | Structures of human peptidylarginine deiminase type III provide insights into substrate recognition and inhibitor design.
Arch.Biochem.Biophys., 708, 2021
|
|
7D5R
 
 | | Structure of the Ca2+-bound C646A mutant of peptidylarginine deiminase type III (PAD3) | | 分子名称: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | 著者 | Mashimo, R, Akimoto, M, Unno, M. | | 登録日 | 2020-09-28 | | 公開日 | 2021-06-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.148 Å) | | 主引用文献 | Structures of human peptidylarginine deiminase type III provide insights into substrate recognition and inhibitor design.
Arch.Biochem.Biophys., 708, 2021
|
|
5HP5
 
 | | Srtucture of human peptidylarginine deiminase type I (PAD1) | | 分子名称: | CALCIUM ION, Protein-arginine deiminase type-1 | | 著者 | Unno, M, Nagai, A, Saijo, S, Shimizu, N, Kinjo, S, Mashimo, R, Kizawa, K, Takahara, H. | | 登録日 | 2016-01-20 | | 公開日 | 2016-07-27 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.198 Å) | | 主引用文献 | Monomeric Form of Peptidylarginine Deiminase Type I Revealed by X-ray Crystallography and Small-Angle X-ray Scattering
J.Mol.Biol., 428, 2016
|
|
7MBJ
 
 | |
7LV3
 
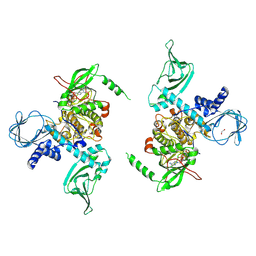 | | Crystal structure of human protein kinase G (PKG) R-C complex in inhibited state | | 分子名称: | 1,2-ETHANEDIOL, Isoform Beta of cGMP-dependent protein kinase 1, MANGANESE (II) ION, ... | | 著者 | Sharma, R, Lying, Q, Casteel, D, Kim, C. | | 登録日 | 2021-02-23 | | 公開日 | 2022-08-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.41 Å) | | 主引用文献 | An auto-inhibited state of protein kinase G and implications for selective activation.
Elife, 11, 2022
|
|
7LZ4
 
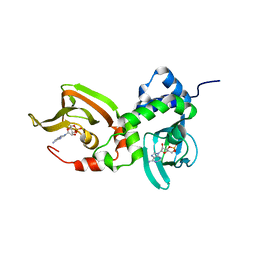 | | Crystal structure of A211D mutant of Protein Kinase A RIa subunit, a Carney Complex mutation | | 分子名称: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, cAMP-dependent protein kinase type I-alpha regulatory subunit, N-terminally processed | | 著者 | Del Rio, J, Wu, J, Taylor, S.S. | | 登録日 | 2021-03-08 | | 公開日 | 2021-05-26 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (4.155 Å) | | 主引用文献 | Noncanonical protein kinase A activation by oligomerization of regulatory subunits as revealed by inherited Carney complex mutations.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7BWI
 
 | | Solution structure of recombinant APETx1 | | 分子名称: | Kappa-actitoxin-Ael2a | | 著者 | Matsumura, K, Kobayashi, N, Kurita, J, Nishimura, Y, Yokogawa, M, Imai, S, Shimada, I, Osawa, M. | | 登録日 | 2020-04-14 | | 公開日 | 2020-12-23 | | 最終更新日 | 2021-07-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Mechanism of hERG inhibition by gating-modifier toxin, APETx1, deduced by functional characterization.
Bmc Mol Cell Biol, 22, 2021
|
|
7DAN
 
 | |
7D8N
 
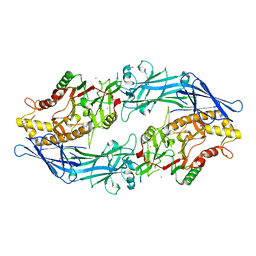 | | Structure of the inactive form of wild-type peptidylarginine deiminase type III (PAD3) crystallized under the condition with high concentrations of Ca2+ | | 分子名称: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | 著者 | Funabashi, K, Sawata, M, Unno, M. | | 登録日 | 2020-10-08 | | 公開日 | 2021-06-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.753 Å) | | 主引用文献 | Structures of human peptidylarginine deiminase type III provide insights into substrate recognition and inhibitor design.
Arch.Biochem.Biophys., 708, 2021
|
|
7D56
 
 | |
7D4Y
 
 | |
