8CN6
 
 | | CD59 in complex with CP-06 peptide | | Descriptor: | CD59 glycoprotein, Cyclic peptide CP-06, ZINC ION | | Authors: | Bickel, J, Ahmed, A, Morgan, M, Horrell, S, El Omari, K, Couves, E.C, Bubeck, D, Tate, E. | | Deposit date: | 2023-02-22 | | Release date: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | A novel cyclic peptide ligand for CD59
To Be Published
|
|
4FYU
 
 | | Crystal structure of Thioredoxin from Wuchereria bancrofti at 2.0 Angstrom | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Thioredoxin | | Authors: | Yousef, N, Prabhu, P, Ahmed, A, Iqbal, S, Betzel, C. | | Deposit date: | 2012-07-05 | | Release date: | 2013-08-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Thioredoxin from Wuchereria bancrofti at 2.0 Angstrom
TO BE PUBLISHED
|
|
8ROY
 
 | | Structure of the human DDB1-DDA1-DCAF15 E3 ubiquitin ligase bound to compound furan 24 | | Descriptor: | 1-[5-[[3,4-bis(chloranyl)-1~{H}-indol-7-yl]sulfamoyl]-3-methyl-furan-2-yl]carbonyl-~{N}-methyl-piperidine-4-carboxamide, DDB1- and CUL4-associated factor 15, DET1- and DDB1-associated protein 1, ... | | Authors: | Shilliday, F, Lucas, S.C.C, Richter, M, Michaelides, I.N, Fusani, L. | | Deposit date: | 2024-01-12 | | Release date: | 2024-04-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Optimization of Potent Ligands for the E3 Ligase DCAF15 and Evaluation of Their Use in Heterobifunctional Degraders.
J.Med.Chem., 67, 2024
|
|
8ROX
 
 | | Structure of the human DDB1-DDA1-DCAF15 E3 ubiquitin ligase bound to compound furan 12 | | Descriptor: | 5-[[3,4-bis(chloranyl)-1~{H}-indol-7-yl]sulfamoyl]-~{N},~{N},3-trimethyl-furan-2-carboxamide;ethane, DDB1- and CUL4-associated factor 15, DET1- and DDB1-associated protein 1, ... | | Authors: | Shilliday, F, Lucas, S.C.C, Richter, M, Michaelides, I.N, Fusani, L. | | Deposit date: | 2024-01-12 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Optimization of Potent Ligands for the E3 Ligase DCAF15 and Evaluation of Their Use in Heterobifunctional Degraders.
J.Med.Chem., 67, 2024
|
|
9EXX
 
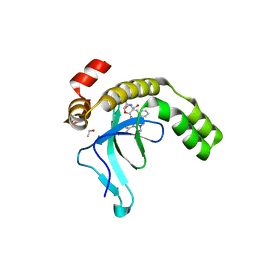 | | Crystal structure of the PWWP1 domain of NSD2 bound by compound 18. | | Descriptor: | 1,2-ETHANEDIOL, 4-methyl-3-[1-methyl-5-(3-oxidanylidene-4~{H}-1,4-benzoxazin-7-yl)imidazol-4-yl]-~{N}-phenyl-benzamide, ETHANOL, ... | | Authors: | Collie, G.W. | | Deposit date: | 2024-04-09 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.943 Å) | | Cite: | Identification of Novel Potent NSD2-PWWP1 Ligands Using Structure-Based Design and Computational Approaches.
J.Med.Chem., 67, 2024
|
|
9EXY
 
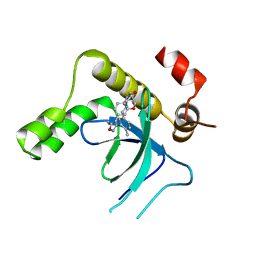 | | Crystal structure of the PWWP1 domain of NSD2 bound by compound 34. | | Descriptor: | 7-[5-methyl-3-[2-methyl-5-(piperidin-1-ylmethyl)phenyl]-1,2-oxazol-4-yl]-4~{H}-1,4-benzoxazin-3-one, Histone-lysine N-methyltransferase NSD2 | | Authors: | Collie, G.W. | | Deposit date: | 2024-04-09 | | Release date: | 2024-05-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Identification of Novel Potent NSD2-PWWP1 Ligands Using Structure-Based Design and Computational Approaches.
J.Med.Chem., 67, 2024
|
|
9EXW
 
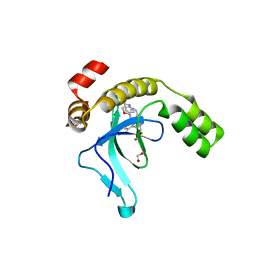 | | Crystal structure of the PWWP1 domain of NSD2 bound by compound 17. | | Descriptor: | 1,2-ETHANEDIOL, 7-[3-methyl-5-[2-methyl-5-[(pyridin-3-ylamino)methyl]phenyl]imidazol-4-yl]-4~{H}-1,4-benzoxazin-3-one, Histone-lysine N-methyltransferase NSD2 | | Authors: | Collie, G.W. | | Deposit date: | 2024-04-09 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Identification of Novel Potent NSD2-PWWP1 Ligands Using Structure-Based Design and Computational Approaches.
J.Med.Chem., 67, 2024
|
|
6UTD
 
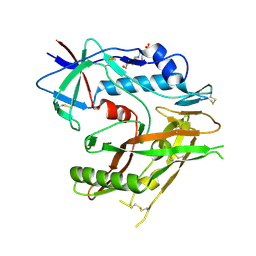 | |
6UT1
 
 | | CRYSTAL STRUCTURE OF HIV-1 LM/HS CLADE A/E CRF01 GP120 CORE IN COMPLEX WITH BNM-III-170 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HIV-1 LM/HS clade A/E CRF01 gp120 core, ... | | Authors: | Tolbert, W.D, Sherburn, R, Pazgier, M. | | Deposit date: | 2019-10-29 | | Release date: | 2020-10-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The HIV-1 Env gp120 Inner Domain Shapes the Phe43 Cavity and the CD4 Binding Site.
Mbio, 11, 2020
|
|
6UTB
 
 | |
6USW
 
 | | CRYSTAL STRUCTURE OF HIV-1 LM/HS CLADE A/E CRF01 GP120 CORE IN COMPLEX WITH (S)-MCG-IV-210 | | Descriptor: | (3S)-N~1~-(2-aminoethyl)-N~3~-(4-chloro-3-fluorophenyl)piperidine-1,3-dicarboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Tolbert, W.D, Sherburn, R, Pazgier, M. | | Deposit date: | 2019-10-28 | | Release date: | 2020-10-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The HIV-1 Env gp120 Inner Domain Shapes the Phe43 Cavity and the CD4 Binding Site.
Mbio, 11, 2020
|
|
7N4I
 
 | |
7N4J
 
 | |
7N4M
 
 | |
7N4L
 
 | |
1SPD
 
 | |
4RQB
 
 | |
4RQA
 
 | |
6XG5
 
 | | X-ray structure of Escherichia coli dihydrofolate reductase in complex with trimethoprim | | Descriptor: | CHLORIDE ION, Dihydrofolate reductase, GLYCEROL, ... | | Authors: | Gaszek, I.K, Manna, M.S, Borek, D, Toprak, E. | | Deposit date: | 2020-06-16 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A trimethoprim derivative impedes antibiotic resistance evolution.
Nat Commun, 12, 2021
|
|
6XG4
 
 | | X-ray structure of Escherichia coli dihydrofolate reductase L28R mutant in complex with trimethoprim | | Descriptor: | CHLORIDE ION, Dihydrofolate reductase, GLYCEROL, ... | | Authors: | Gaszek, I.K, Manna, M.S, Borek, D, Toprak, E. | | Deposit date: | 2020-06-16 | | Release date: | 2021-03-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A trimethoprim derivative impedes antibiotic resistance evolution.
Nat Commun, 12, 2021
|
|
4E52
 
 | |
5BVB
 
 | | Engineered Digoxigenin binder DIG5.1a | | Descriptor: | DIG5.1a, DIGOXIGENIN | | Authors: | Doyle, L.A, Stoddard, B.L. | | Deposit date: | 2015-06-04 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | CSAR Benchmark Exercise 2013: Evaluation of Results from a Combined Computational Protein Design, Docking, and Scoring/Ranking Challenge.
J.Chem.Inf.Model., 56, 2016
|
|
4HPS
 
 | |
4HKM
 
 | |
5OXS
 
 | |
