4ENF
 
 | | Crystal structure of the cap-binding domain of polymerase basic protein 2 from influenza virus A/Puerto Rico/8/34(h1n1) | | Descriptor: | 1,4-BUTANEDIOL, NITRATE ION, Polymerase basic protein 2 | | Authors: | Meng, G, Liu, Y, Zheng, X. | | Deposit date: | 2012-04-13 | | Release date: | 2013-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structural and functional characterization of K339T substitution identified in the PB2 subunit cap-binding pocket of influenza A virus
J.Biol.Chem., 288, 2013
|
|
4ES5
 
 | |
1ON3
 
 | | Transcarboxylase 12S crystal structure: hexamer assembly and substrate binding to a multienzyme core (with methylmalonyl-coenzyme a and methylmalonic acid bound) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CADMIUM ION, METHYLMALONIC ACID, ... | | Authors: | Hall, P.R, Wang, Y.-F, Rivera-Hainaj, R.E, Zheng, X, Pustai-Carey, M, Carey, P.R, Yee, V.C. | | Deposit date: | 2003-02-26 | | Release date: | 2003-05-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Transcarboxylase 12S crystal structure: hexamer assembly and substrate binding to a multienzyme core
Embo J., 22, 2003
|
|
2XPK
 
 | | Cell-penetrant, nanomolar O-GlcNAcase inhibitors selective against lysosomal hexosaminidases | | Descriptor: | N-[(5R,6R,7R,8S)-6,7-DIHYDROXY-5-(HYDROXYMETHYL)-2-(2-PHENYLETHYL)-5,6,7,8-TETRAHYDROIMIDAZO[1,2-A]PYRIDIN-8-YL]-3-SULFANYLPROPANAMIDE, O-GLCNACASE NAGJ | | Authors: | Dorfmueller, H.C, Borodkin, V.S, Schimpl, M, Zheng, X, Kime, R, Read, K.D, van Aalten, D.M.F. | | Deposit date: | 2010-08-26 | | Release date: | 2011-03-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Cell-Penetrant, Nanomolar O-Glcnacase Inhibitors Selective Against Lysosomal Hexosaminidases.
Chem.Biol, 17, 2010
|
|
1ON9
 
 | | Transcarboxylase 12S crystal structure: hexamer assembly and substrate binding to a multienzyme core (with hydrolyzed methylmalonyl-coenzyme a bound) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CADMIUM ION, METHYLMALONYL-COENZYME A, ... | | Authors: | Hall, P.R, Wang, Y.-F, Rivera-Hainaj, R.E, Zheng, X, Pustai-Carey, M, Carey, P.R, Yee, V.C. | | Deposit date: | 2003-02-27 | | Release date: | 2003-05-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Transcarboxylase 12S crystal structure: hexamer assembly and substrate binding to a multienzyme core
Embo J., 22, 2003
|
|
3KH8
 
 | | Crystal structure of MaoC-like dehydratase from Phytophthora Capsici | | Descriptor: | MaoC-like dehydratase | | Authors: | Wang, H, Zhang, K, Guo, J, Zhou, Q, Zheng, X, Sun, F, Pang, H, Zhang, X. | | Deposit date: | 2009-10-30 | | Release date: | 2010-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of MaoC-like dehydratase from Phytophthora Capsici
To be Published
|
|
6K8N
 
 | | Crystal structure of the Sulfolobus solfataricus topoisomerase III | | Descriptor: | ZINC ION, topoisomerase III | | Authors: | Wang, H.Q, Zhang, J.H, Zheng, X, Zheng, Z.F, Dong, Y.H, Huang, L, Gong, Y. | | Deposit date: | 2019-06-13 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the Sulfolobus solfataricus topoisomerase III reveal that its C-terminal novel zinc finger part is a unique decatenation domain
To Be Published
|
|
6K8O
 
 | | Crystal structure of the Sulfolobus solfataricus topoisomerase III in complex with DNA | | Descriptor: | DNA (5'-D(*GP*CP*AP*AP*GP*GP*TP*C)-3'), ZINC ION, topoisomerase III | | Authors: | Wang, H.Q, Zhang, J.H, Zheng, X, Zheng, Z.F, Dong, Y.H, Huang, L, Gong, Y. | | Deposit date: | 2019-06-13 | | Release date: | 2020-06-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of the Sulfolobus solfataricus topoisomerase III reveal that its C-terminal novel zinc finger part is a unique decatenation domain
To Be Published
|
|
3E5M
 
 | | Crystal structure of the HSCARG Y81A mutant | | Descriptor: | NmrA-like family domain-containing protein 1 | | Authors: | Li, Y, Meng, G, Dai, X, Luo, M, Zheng, X. | | Deposit date: | 2008-08-14 | | Release date: | 2009-05-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | NADPH is an allosteric regulator of HSCARG
J.Mol.Biol., 387, 2009
|
|
3DXF
 
 | | Crystal structure of the HSCARG R37A mutant | | Descriptor: | NmrA-like family domain-containing protein 1 | | Authors: | Li, Y, Meng, G, Dai, X, Luo, M, Zheng, X. | | Deposit date: | 2008-07-24 | | Release date: | 2009-05-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | NADPH is an allosteric regulator of HSCARG
J.Mol.Biol., 387, 2009
|
|
4YVV
 
 | | Crystal structure of AKR1C3 complexed with glibenclamide | | Descriptor: | 5-chloro-N-(2-{4-[(cyclohexylcarbamoyl)sulfamoyl]phenyl}ethyl)-2-methoxybenzamide, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhao, Y, Zheng, X, Zhang, H, Hu, X. | | Deposit date: | 2015-03-20 | | Release date: | 2015-11-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | In vitro inhibition of AKR1Cs by sulphonylureas and the structural basis
Chem.Biol.Interact., 240, 2015
|
|
4YVX
 
 | | Crystal structure of AKR1C3 complexed with glimepiride | | Descriptor: | 3-ethyl-4-methyl-N-[2-(4-{[(cis-4-methylcyclohexyl)carbamoyl]sulfamoyl}phenyl)ethyl]-2-oxo-2,5-dihydro-1H-pyrrole-1-car boxamide, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhao, Y, Zheng, X, Zhang, H, Hu, X. | | Deposit date: | 2015-03-20 | | Release date: | 2015-11-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | In vitro inhibition of AKR1Cs by sulphonylureas and the structural basis
Chem.Biol.Interact., 240, 2015
|
|
4YVP
 
 | | Crystal Structure of AKR1C1 complexed with glibenclamide | | Descriptor: | 5-chloro-N-(2-{4-[(cyclohexylcarbamoyl)sulfamoyl]phenyl}ethyl)-2-methoxybenzamide, Aldo-keto reductase family 1 member C1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhao, Y, Zheng, X, Zhang, H, Hu, X. | | Deposit date: | 2015-03-20 | | Release date: | 2015-11-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | In vitro inhibition of AKR1Cs by sulphonylureas and the structural basis
Chem.Biol.Interact., 240, 2015
|
|
3QNI
 
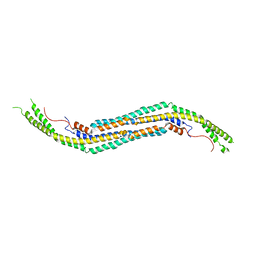 | |
3RFE
 
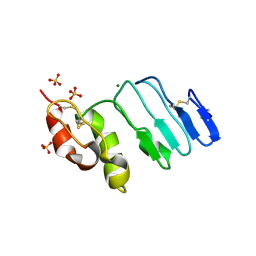 | | Crystal structure of glycoprotein GPIb ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Platelet glycoprotein Ib beta chain, ... | | Authors: | McEwan, P.A, Yang, W, Carr, K.H, Mo, X, Zheng, X, Li, R, Emsley, J. | | Deposit date: | 2011-04-06 | | Release date: | 2011-12-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.245 Å) | | Cite: | Quaternary organization of GPIb-IX complex and insights into Bernard-Soulier syndrome revealed by the structures of GPIbbeta and a GPIbbeta/GPIX chimer
Blood, 118, 2011
|
|
3REZ
 
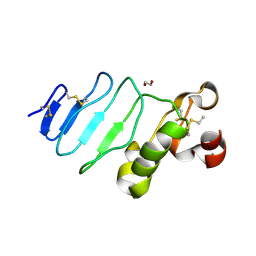 | | glycoprotein GPIb variant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, Platelet glycoprotein Ib beta chain, ... | | Authors: | McEwan, P.A, Yang, W, Carr, K.H, Mo, X, Zheng, X, Li, R, Emsley, J. | | Deposit date: | 2011-04-05 | | Release date: | 2012-02-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Quaternary organization of GPIb-IX complex and insights into Bernard-Soulier syndrome revealed by the structures of GPIb beta and a GPIb beta/GPIX chimera
Blood, 118, 2011
|
|
3MX7
 
 | |
3QE6
 
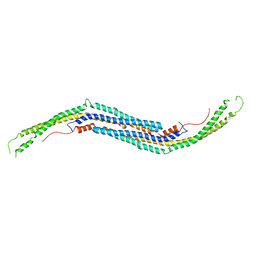 | | Mouse PACSIN 3 F-BAR domain structure | | Descriptor: | MAGNESIUM ION, Protein kinase C and casein kinase II substrate protein 3 | | Authors: | Meng, G, Bai, X, Zheng, X. | | Deposit date: | 2011-01-19 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Rigidity of wedge loop in PACSIN 3 protein is a key factor in dictating diameters of tubules
J.Biol.Chem., 287, 2012
|
|
3Q0K
 
 | | Crystal structure of Human PACSIN 2 F-BAR | | Descriptor: | CALCIUM ION, Protein kinase C and casein kinase substrate in neurons protein 2 | | Authors: | Bai, X, Meng, G, Zheng, X. | | Deposit date: | 2010-12-15 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of human PACSIN 2 F-BAR domain
To be Published
|
|
4H5Y
 
 | | High-resolution crystal structure of Legionella pneumophila LidA (60-594) | | Descriptor: | LidA protein, substrate of the Dot/Icm system | | Authors: | An, X, Ye, S, Liu, Y, Zheng, X, Zhang, R. | | Deposit date: | 2012-09-19 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of LidA, a translocated substrate of the Legionella pneumophila type IV secretion system.
Protein Cell, 4, 2013
|
|
4GQG
 
 | | Crystal structure of AKR1B10 complexed with NADP+ | | Descriptor: | Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhang, L, Zheng, X, Chen, S, Zhai, J, Hu, X. | | Deposit date: | 2012-08-23 | | Release date: | 2013-08-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Inhibitor selectivity between aldo-keto reductase superfamily members AKR1B10 and AKR1B1: Role of Trp112 (Trp111)
Febs Lett., 587, 2013
|
|
5JZV
 
 | | The structure of D77G hCINAP-ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Adenylate kinase isoenzyme 6 | | Authors: | Liu, Y, Yang, Z, Yang, Y, Cai, X, Zheng, X. | | Deposit date: | 2016-05-17 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | The ATPase hCINAP regulates 18S rRNA processing and is essential for embryogenesis and tumour growth.
Nat Commun, 7, 2016
|
|
4I5X
 
 | | Crystal Structure Of AKR1B10 Complexed With NADP+ And Flufenamic acid | | Descriptor: | 2-[[3-(TRIFLUOROMETHYL)PHENYL]AMINO] BENZOIC ACID, Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhang, L, Zheng, X, Chen, S, Zhai, J, Zhang, H, Zhao, Y. | | Deposit date: | 2012-11-29 | | Release date: | 2013-10-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibitor selectivity between aldo-keto reductase superfamily members AKR1B10 and AKR1B1: Role of Trp112 (Trp111)
Febs Lett., 587, 2013
|
|
4N9B
 
 | | Fragment-based Design of 3-Aminopyridine-derived Amides as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | Descriptor: | 1-methyl-N-(pyridin-3-yl)-1H-pyrazole-5-carboxamide, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Dragovich, P.S, Zhao, G, Baumeister, T, Bravo, B, Giannetti, A.M, Ho, Y, Hua, R, Li, G, Liang, X, O'Brien, T, Skelton, N.J, Wang, C, Zhai, Q, Oh, A, Wang, W, Wang, Y, Xiao, Y, Yuen, P, Zak, M, Zheng, X. | | Deposit date: | 2013-10-20 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.859 Å) | | Cite: | Fragment-based design of 3-aminopyridine-derived amides as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4N9D
 
 | | Fragment-based Design of 3-Aminopyridine-derived Amides as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | Descriptor: | 1,2-ETHANEDIOL, 4-({[(4-tert-butylphenyl)sulfonyl]amino}methyl)-N-(pyridin-3-yl)benzamide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Dragovich, P.S, Zhao, G, Baumeister, T, Bravo, B, Giannetti, A.M, Ho, Y, Hua, R, Li, G, Liang, X, O'Brien, T, Skelton, N.J, Wang, C, Zhao, Q, Oh, A, Wang, W, Wang, Y, Xiao, Y, Yuen, P, Zak, M, Zheng, X. | | Deposit date: | 2013-10-20 | | Release date: | 2014-02-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Fragment-based design of 3-aminopyridine-derived amides as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 24, 2014
|
|
