6D2L
 
 | | Crystal structure of human CARM1 with (S)-SKI-72 | | 分子名称: | (2S,5S)-2-amino-6-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]-5-[(benzylamino)methyl]-N-[2-(4-hydroxyphenyl)ethyl]hexanamide, GLYCEROL, Histone-arginine methyltransferase CARM1, ... | | 著者 | DONG, A, ZENG, H, WALKER, J.R, Hutchinson, A, Seitova, A, LUO, M, CAI, X.C, KE, W, WANG, J, SHI, C, ZHENG, W, LEE, J.P, IBANEZ, G, Bountra, C, Arrowsmith, C.H, Edwards, A.M, BROWN, P.J, WU, H, Structural Genomics Consortium (SGC) | | 登録日 | 2018-04-13 | | 公開日 | 2018-05-23 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | A chemical probe of CARM1 alters epigenetic plasticity against breast cancer cell invasion.
Elife, 8, 2019
|
|
7CQQ
 
 | | GmaS in complex with AMPPNP and MetSox | | 分子名称: | (2S)-2-AMINO-4-(METHYLSULFONIMIDOYL)BUTANOIC ACID, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | 著者 | Li, C.Y, Zhang, Y.Z. | | 登録日 | 2020-08-11 | | 公開日 | 2020-11-18 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.295 Å) | | 主引用文献 | Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine.
J.Biol.Chem., 296, 2020
|
|
7CQL
 
 | | Apo GmaS without ligand | | 分子名称: | Type III glutamate--ammonia ligase | | 著者 | Li, C.Y, Zhang, Y.Z. | | 登録日 | 2020-08-11 | | 公開日 | 2020-11-18 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.801 Å) | | 主引用文献 | Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine.
J.Biol.Chem., 296, 2020
|
|
7CQN
 
 | | GmaS in complex with AMPPCP | | 分子名称: | PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Type III glutamate--ammonia ligase | | 著者 | Li, C.Y, Zhang, Y.Z. | | 登録日 | 2020-08-11 | | 公開日 | 2020-11-18 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.962 Å) | | 主引用文献 | Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine.
J.Biol.Chem., 296, 2020
|
|
7CQU
 
 | | GmaS/ADP/MetSox-P complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, L-METHIONINE-S-SULFOXIMINE PHOSPHATE, MAGNESIUM ION, ... | | 著者 | Li, C.Y, Zhang, Y.Z. | | 登録日 | 2020-08-11 | | 公開日 | 2020-11-18 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine.
J.Biol.Chem., 296, 2020
|
|
7CQX
 
 | | GmaS/ADP complex-Conformation 2 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Type III glutamate--ammonia ligase | | 著者 | Li, C.Y, Zhang, Y.Z. | | 登録日 | 2020-08-11 | | 公開日 | 2020-11-18 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.301 Å) | | 主引用文献 | Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine.
J.Biol.Chem., 296, 2020
|
|
7CQW
 
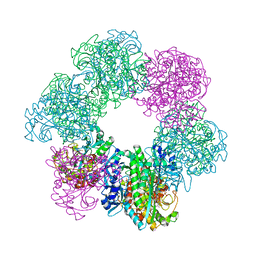 | | GmaS/ADP complex-Conformation 1 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Type III glutamate--ammonia ligase | | 著者 | Li, C.Y, Zhang, Y.Z. | | 登録日 | 2020-08-11 | | 公開日 | 2020-11-18 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.297 Å) | | 主引用文献 | Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine.
J.Biol.Chem., 296, 2020
|
|
6K7O
 
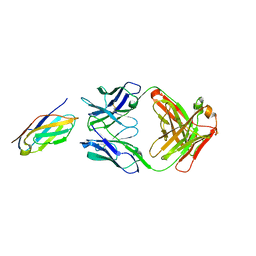 | | Complex structure of LILRB4 and h128-3 antibody | | 分子名称: | Leukocyte immunoglobulin-like receptor subfamily B member 4, h128-3 Fab heavy chain, h128-3 Fab light chain | | 著者 | Song, H, Chai, Y, Xu, X, Gao, F.G. | | 登録日 | 2019-06-08 | | 公開日 | 2019-07-03 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.004 Å) | | 主引用文献 | Disrupting LILRB4/APOE Interaction by an Efficacious Humanized Antibody Reverses T-cell Suppression and Blocks AML Development.
Cancer Immunol Res, 7, 2019
|
|
6UPL
 
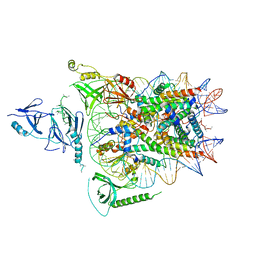 | | Structure of FACT_subnucleosome complex 2 | | 分子名称: | DNA (79-mer), FACT complex subunit SPT16, FACT complex subunit SSRP1, ... | | 著者 | Zhou, K, Tan, Y.Z, Wei, H, Liu, Y, Carragher, B, Potter, C, Luger, K. | | 登録日 | 2019-10-17 | | 公開日 | 2019-12-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (7.4 Å) | | 主引用文献 | FACT caught in the act of manipulating the nucleosome.
Nature, 577, 2020
|
|
6UPK
 
 | | Structure of FACT_subnucleosome complex 1 | | 分子名称: | DNA (79-mer), FACT complex subunit SPT16, FACT complex subunit SSRP1, ... | | 著者 | Zhou, K, Tan, Y.Z, Wei, H, Liu, Y, Carragher, B, Potter, C, Luger, K. | | 登録日 | 2019-10-17 | | 公開日 | 2019-12-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.9 Å) | | 主引用文献 | FACT caught in the act of manipulating the nucleosome.
Nature, 577, 2020
|
|
7KXT
 
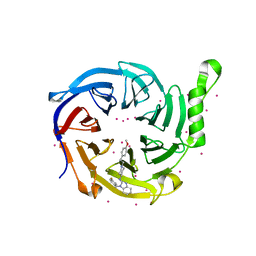 | | Crystal structure of human EED | | 分子名称: | 1-[(4-fluorophenyl)methyl]-N-{1-[2-(4-methoxyphenyl)ethyl]piperidin-4-yl}-1H-benzimidazol-2-amine, Polycomb protein EED, UNKNOWN ATOM OR ION | | 著者 | Zhu, L, Dong, A, Du, D, Liu, Y, Luo, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2020-12-04 | | 公開日 | 2021-02-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Structure-Guided Development of Small-Molecule PRC2 Inhibitors Targeting EZH2-EED Interaction.
J.Med.Chem., 64, 2021
|
|
1FB9
 
 | |
7M1C
 
 | |
7M30
 
 | |
7LYV
 
 | |
7M22
 
 | |
7LYW
 
 | |
6ILG
 
 | |
6ILF
 
 | |
6ILC
 
 | |
6ILE
 
 | | CRYSTAL STRUCTURE OF A MUTANT PTAL-N*01:01 FOR 2.9 ANGSTROM, 52M 53D 54L DELETED | | 分子名称: | Beta-2-microglobulin, HEV-1, MHC class I antigen | | 著者 | Qu, Z.H, Zhang, N.Z, Xia, C. | | 登録日 | 2018-10-17 | | 公開日 | 2019-07-24 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structure and Peptidome of the Bat MHC Class I Molecule Reveal a Novel Mechanism Leading to High-Affinity Peptide Binding.
J Immunol., 202, 2019
|
|
6KX2
 
 | | Crystal structure of GDP bound RhoA protein | | 分子名称: | GUANOSINE-5'-DIPHOSPHATE, Transforming protein RhoA | | 著者 | Zhang, H, Luo, C. | | 登録日 | 2019-09-09 | | 公開日 | 2020-08-19 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.454 Å) | | 主引用文献 | Covalent Inhibitors Allosterically Block the Activation of Rho Family Proteins and Suppress Cancer Cell Invasion.
Adv Sci, 7, 2020
|
|
6KX3
 
 | | Crystal structure of RhoA protein with covalent inhibitor DC-Rhoin | | 分子名称: | GUANOSINE-5'-DIPHOSPHATE, Transforming protein RhoA, prop-2-enyl (3R)-1,1-bis(oxidanylidene)-2,3-dihydro-1-benzothiophene-3-carboxylate | | 著者 | Zhang, H, Luo, C. | | 登録日 | 2019-09-09 | | 公開日 | 2020-08-19 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.981 Å) | | 主引用文献 | Covalent Inhibitors Allosterically Block the Activation of Rho Family Proteins and Suppress Cancer Cell Invasion.
Adv Sci, 7, 2020
|
|
6IRL
 
 | |
6WYV
 
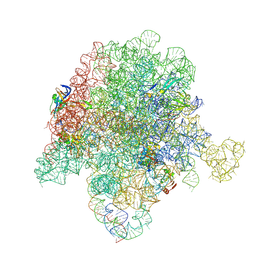 | | E. coli 50S ribosome bound to compounds 47 and VS1 | | 分子名称: | (3R,4R,5E,10E,12E,14S,16R,26aR)-16-fluoro-14-hydroxy-12-methyl-3-(propan-2-yl)-4-(prop-2-en-1-yl)-3,4,8,9,14,15,16,17,24,25,26,26a-dodecahydro-1H,7H,22H-21,18-(azeno)pyrrolo[2,1-c][1,8,4,19]dioxadiazacyclotetracosine-1,7,22-trione, 23S ribosomal RNA, 50S ribosomal protein L13, ... | | 著者 | Pellegrino, J, Lee, D.J, Fraser, J.S, Seiple, I.B. | | 登録日 | 2020-05-13 | | 公開日 | 2020-06-17 | | 最終更新日 | 2020-10-07 | | 実験手法 | ELECTRON MICROSCOPY (2.75 Å) | | 主引用文献 | Synthetic group A streptogramin antibiotics that overcome Vat resistance.
Nature, 586, 2020
|
|
