7E5X
 
 | |
2JXB
 
 | | Structure of CD3epsilon-Nck2 first SH3 domain complex | | 分子名称: | T-cell surface glycoprotein CD3 epsilon chain, Cytoplasmic protein NCK2 | | 著者 | Takeuchi, K, Yang, H, Ng, E, Park, S, Sun, Z.J, Reinherz, E.L, Wagner, G. | | 登録日 | 2007-11-09 | | 公開日 | 2008-09-23 | | 最終更新日 | 2022-03-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural and functional evidence that Nck interaction with CD3epsilon regulates T-cell receptor activity.
J.Mol.Biol., 380, 2008
|
|
7VUX
 
 | | Complex structure of PD1 and 609A-Fab | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Huang, H, Zhu, Z, Zhao, J, Jiang, L, Yang, H, Deng, L, Meng, X, Ding, J, Yang, S, Zhao, L, Xu, W, Wang, X. | | 登録日 | 2021-11-04 | | 公開日 | 2021-11-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | A strategy for the efficient construction of anti-PD1-based bispecific antibodies with desired IgG-like properties.
Mabs, 14, 2022
|
|
7E3L
 
 | | Ultrapotent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 58G6 heavy chain, 58G6 light chain, ... | | 著者 | Guo, H, Li, T, Liu, F, Gao, Y, Ji, X, Yang, H. | | 登録日 | 2021-02-09 | | 公開日 | 2021-09-15 | | 最終更新日 | 2022-03-02 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Potent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants.
Nat Commun, 12, 2021
|
|
7E3K
 
 | | Ultrapotent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants | | 分子名称: | 13G9 heavy chain, 13G9 light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Guo, H, Li, T, Liu, F, Gao, Y, Ji, X, Yang, H. | | 登録日 | 2021-02-09 | | 公開日 | 2021-09-15 | | 最終更新日 | 2022-03-02 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Potent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants.
Nat Commun, 12, 2021
|
|
6LND
 
 | |
6LNB
 
 | | CryoEM structure of Cascade-TniQ-dsDNA complex | | 分子名称: | CRISPR RNA (60-MER), CRISPR-associated protein Cas6, CRISPR-associated protein Cas7, ... | | 著者 | Wang, B, Xu, W, Yang, H. | | 登録日 | 2019-12-28 | | 公開日 | 2020-02-19 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.18 Å) | | 主引用文献 | Structural basis of a Tn7-like transposase recruitment and DNA loading to CRISPR-Cas surveillance complex.
Cell Res., 30, 2020
|
|
6LNC
 
 | | CryoEM structure of Cascade-TniQ complex | | 分子名称: | CRISPR RNA (60-MER), CRISPR-associated protein Cas6, CRISPR-associated protein Cas7, ... | | 著者 | Wang, B, Xu, W, Yang, H. | | 登録日 | 2019-12-28 | | 公開日 | 2020-02-19 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.21 Å) | | 主引用文献 | Structural basis of a Tn7-like transposase recruitment and DNA loading to CRISPR-Cas surveillance complex.
Cell Res., 30, 2020
|
|
6NR3
 
 | | Cryo-EM structure of the TRPM8 ion channel in complex with high occupancy icilin, PI(4,5)P2, and calcium | | 分子名称: | (2S)-1-{[(R)-hydroxy{[(1R,2R,3S,4R,5R,6S)-2,3,6-trihydroxy-4,5-bis(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}-3-(octadecanoyloxy)propan-2-yl icosa-5,8,11,14-tetraenoate, CALCIUM ION, Icilin, ... | | 著者 | Yin, Y, Le, S.C, Hsu, A.L, Borgnia, M.J, Yang, H, Lee, S.-Y. | | 登録日 | 2019-01-22 | | 公開日 | 2019-02-20 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural basis of cooling agent and lipid sensing by the cold-activated TRPM8 channel.
Science, 363, 2019
|
|
6NR4
 
 | | Cryo-EM structure of the TRPM8 ion channel with low occupancy icilin, PI(4,5)P2, and calcium | | 分子名称: | Transient receptor potential cation channel subfamily M member 8 | | 著者 | Yin, Y, Le, S.C, Hsu, A.L, Borgnia, M.J, Yang, H, Lee, S.-Y. | | 登録日 | 2019-01-22 | | 公開日 | 2019-02-20 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Structural basis of cooling agent and lipid sensing by the cold-activated TRPM8 channel.
Science, 363, 2019
|
|
6NR2
 
 | | Cryo-EM structure of the TRPM8 ion channel in complex with the menthol analog WS-12 and PI(4,5)P2 | | 分子名称: | (1R,2S,5R)-N-(4-methoxyphenyl)-5-methyl-2-(propan-2-yl)cyclohexane-1-carboxamide, (2S)-1-{[(R)-hydroxy{[(1R,2R,3S,4R,5R,6S)-2,3,6-trihydroxy-4,5-bis(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}-3-(octadecanoyloxy)propan-2-yl icosa-5,8,11,14-tetraenoate, Transient receptor potential cation channel subfamily M member 8 | | 著者 | Yin, Y, Le, S.C, Hsu, A.L, Borgnia, M.J, Yang, H, Lee, S.-Y. | | 登録日 | 2019-01-22 | | 公開日 | 2019-02-20 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Structural basis of cooling agent and lipid sensing by the cold-activated TRPM8 channel.
Science, 363, 2019
|
|
5XQ1
 
 | | Structural basis of kindlin-mediated integrin recognition and activation | | 分子名称: | Fermitin family homolog 2,Integrin beta-3 | | 著者 | Li, H, Yang, H, Sun, K, Zhang, Z, Yu, C, Wei, Z. | | 登録日 | 2017-06-05 | | 公開日 | 2017-07-26 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.954 Å) | | 主引用文献 | Structural basis of kindlin-mediated integrin recognition and activation
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5XPY
 
 | | Structural basis of kindlin-mediated integrin recognition and activation | | 分子名称: | ACETATE ION, Fermitin family homolog 2, GLYCEROL | | 著者 | Li, H, Yang, H, Sun, K, Zhang, Z, Yu, C, Wei, Z. | | 登録日 | 2017-06-05 | | 公開日 | 2017-07-26 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.099 Å) | | 主引用文献 | Structural basis of kindlin-mediated integrin recognition and activation
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5XPZ
 
 | | Structural basis of kindlin-mediated integrin recognition and activation | | 分子名称: | Fermitin family homolog 2, GLYCEROL | | 著者 | Li, H, Yang, H, Sun, K, Zhang, Z, Yu, C, Wei, Z. | | 登録日 | 2017-06-05 | | 公開日 | 2017-07-26 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.601 Å) | | 主引用文献 | Structural basis of kindlin-mediated integrin recognition and activation
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5XQ0
 
 | | Structural basis of kindlin-mediated integrin recognition and activation | | 分子名称: | Fermitin family homolog 2,Integrin beta-1, GLYCEROL | | 著者 | Li, H, Yang, H, Sun, K, Zhang, Z, Yu, C, Wei, Z. | | 登録日 | 2017-06-05 | | 公開日 | 2017-07-26 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Structural basis of kindlin-mediated integrin recognition and activation
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5ZHZ
 
 | | Crystal structure of the apurinic/apyrimidinic endonuclease IV from Mycobacterium tuberculosis | | 分子名称: | Probable endonuclease 4, SULFATE ION, ZINC ION | | 著者 | Zhang, W, Xu, Y, Yan, M, Li, S, Wang, H, Yang, H, Zhou, W, Rao, Z. | | 登録日 | 2018-03-13 | | 公開日 | 2018-04-04 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.18 Å) | | 主引用文献 | Crystal structure of the apurinic/apyrimidinic endonuclease IV from Mycobacterium tuberculosis.
Biochem. Biophys. Res. Commun., 498, 2018
|
|
7VAH
 
 | |
7WIX
 
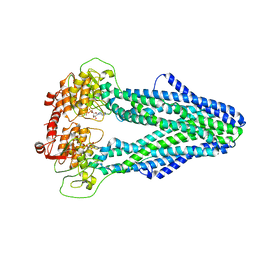 | | Cryo-EM structure of Mycobacterium tuberculosis irtAB in complex with ADP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Mycobactin import ATP-binding/permease protein IrtA, ... | | 著者 | Zhang, B, Sun, S, Yang, H, Rao, Z. | | 登録日 | 2022-01-05 | | 公開日 | 2023-01-25 | | 最終更新日 | 2024-04-10 | | 実験手法 | ELECTRON MICROSCOPY (3.53 Å) | | 主引用文献 | Cryo-EM structures for the Mycobacterium tuberculosis iron-loaded siderophore transporter IrtAB.
Protein Cell, 14, 2023
|
|
7WIV
 
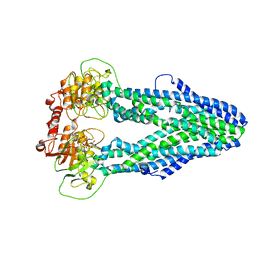 | | Cryo-EM structure of Mycobacterium tuberculosis irtAB in complex with an AMP-PNP | | 分子名称: | Mycobactin import ATP-binding/permease protein IrtA, Mycobactin import ATP-binding/permease protein IrtB, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | 著者 | Zhang, B, Sun, S, Yang, H, Rao, Z. | | 登録日 | 2022-01-05 | | 公開日 | 2023-01-25 | | 最終更新日 | 2024-04-10 | | 実験手法 | ELECTRON MICROSCOPY (2.88 Å) | | 主引用文献 | Cryo-EM structures for the Mycobacterium tuberculosis iron-loaded siderophore transporter IrtAB.
Protein Cell, 14, 2023
|
|
7WIU
 
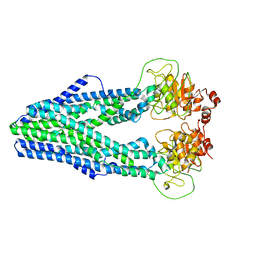 | | Cryo-EM structure of Mycobacterium tuberculosis irtAB in inward-facing state | | 分子名称: | Mycobactin import ATP-binding/permease protein IrtA, Mycobactin import ATP-binding/permease protein IrtB | | 著者 | Zhang, B, Sun, S, Yang, H, Rao, Z. | | 登録日 | 2022-01-05 | | 公開日 | 2023-01-25 | | 最終更新日 | 2024-04-10 | | 実験手法 | ELECTRON MICROSCOPY (3.48 Å) | | 主引用文献 | Cryo-EM structures for the Mycobacterium tuberculosis iron-loaded siderophore transporter IrtAB.
Protein Cell, 14, 2023
|
|
7WIW
 
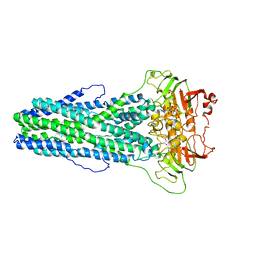 | | Cryo-EM structure of Mycobacterium tuberculosis irtAB complexed with ATP in an occluded conformation | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Mycobactin import ATP-binding/permease protein IrtA, ... | | 著者 | Zhang, B, Sun, S, Yang, H, Rao, Z. | | 登録日 | 2022-01-05 | | 公開日 | 2023-01-25 | | 最終更新日 | 2024-04-10 | | 実験手法 | ELECTRON MICROSCOPY (3.12 Å) | | 主引用文献 | Cryo-EM structures for the Mycobacterium tuberculosis iron-loaded siderophore transporter IrtAB.
Protein Cell, 14, 2023
|
|
7WWK
 
 | | Local refinement of the SARS-CoV-2 BA.1 Spike trimer in complex with 55A8 Fab | | 分子名称: | 55A8 light chain, Spike glycoprotein | | 著者 | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | 登録日 | 2022-02-13 | | 公開日 | 2023-02-15 | | 最終更新日 | 2023-04-19 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Local refinement of the SARS-CoV-2 BA.1 Spike trimer in complex with 55A8 Fab
To Be Published
|
|
7WWI
 
 | | SARS-CoV-2 BA.1 Spike trimer in complex with 55A8 Fab in the class 1 conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 55A8 heavy chain, ... | | 著者 | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | 登録日 | 2022-02-13 | | 公開日 | 2023-02-15 | | 最終更新日 | 2023-04-19 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | SARS-CoV-2 BA.1 Spike trimer in complex with 55A8 Fab in the class 1 conformation
To Be Published
|
|
7WWJ
 
 | | SARS-CoV-2 BA.1 Spike trimer in complex with 55A8 Fab in the class 2 conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 55A8 heavy chain, ... | | 著者 | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | 登録日 | 2022-02-13 | | 公開日 | 2023-02-15 | | 最終更新日 | 2023-04-19 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | EM structure of SARS-CoV-2 Omicron variant spike glycoprotein and 55A8
To Be Published
|
|
7XJ6
 
 | | SARS-CoV-2 BA.1 Spike trimer in complex with 55A8 Fab and 58G6 Fab in the class 1 conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 55A8 heavy chain, ... | | 著者 | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | 登録日 | 2022-04-15 | | 公開日 | 2023-04-19 | | 実験手法 | ELECTRON MICROSCOPY (3.29 Å) | | 主引用文献 | SARS-CoV-2 BA.1 Spike trimer in complex with 55A8 Fab and 58G6 Fab in the class 1 conformation
To Be Published
|
|
