5B4J
 
 | | Crystal structure of I86D mutant of phycocyanobilin:ferredoxin oxidoreductase in complex with biliverdin (data 3) | | Descriptor: | BILIVERDINE IX ALPHA, Phycocyanobilin:ferredoxin oxidoreductase | | Authors: | Hagiwara, Y, Wada, K, Irikawa, T, Unno, M, Fukuyama, K, Sugishima, M. | | Deposit date: | 2016-04-04 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Atomic-resolution structure of the phycocyanobilin:ferredoxin oxidoreductase I86D mutant in complex with fully protonated biliverdin
FEBS Lett., 590, 2016
|
|
7V4N
 
 | |
7V4O
 
 | | Unique non-heme hydroxylase in biosynthesis of nucleoside antibiotic pathway uncover mechanism of reaction | | Descriptor: | 3,6,9,12,15-PENTAOXAHEPTADECANE, Beta-hydroxylase | | Authors: | Li, T.L, Saeid, M.Z. | | Deposit date: | 2021-08-13 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | beta-Hydroxylation of alpha-amino-beta-hydroxylbutanoyl-glycyluridine catalyzed by a nonheme hydroxylase ensures the maturation of caprazamycin
Commun Chem, 5, 2022
|
|
7V4M
 
 | |
7V4F
 
 | |
7V4P
 
 | |
2K86
 
 | | Solution Structure of FOXO3a Forkhead domain | | Descriptor: | Forkhead box protein O3 | | Authors: | Wang, F, Marshall, C.B, Li, G, Plevin, M.J, Ikura, M. | | Deposit date: | 2008-09-02 | | Release date: | 2008-10-14 | | Last modified: | 2020-02-19 | | Method: | SOLUTION NMR | | Cite: | Biochemical and structural characterization of an intramolecular interaction in FOXO3a and its binding with p53.
J.Mol.Biol., 384, 2008
|
|
1BMD
 
 | |
3UMV
 
 | | Eukaryotic Class II CPD photolyase structure reveals a basis for improved UV-tolerance in plants | | Descriptor: | 1,2-ETHANEDIOL, Deoxyribodipyrimidine photo-lyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Arvai, A.S, Hitomi, K, Getzoff, E.D, Tainer, J.A. | | Deposit date: | 2011-11-14 | | Release date: | 2011-12-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.705 Å) | | Cite: | Eukaryotic Class II Cyclobutane Pyrimidine Dimer Photolyase Structure Reveals Basis for Improved Ultraviolet Tolerance in Plants.
J.Biol.Chem., 287, 2012
|
|
5BXT
 
 | | LNBase in complex with LNB-NHAcAUS | | Descriptor: | Lacto-N-biosidase, N-{[(1R,2R,3R,7S,7aR)-1,2,7-trihydroxyhexahydro-1H-pyrrolizin-3-yl]methyl}acetamide, SULFATE ION, ... | | Authors: | Ito, T, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-06-09 | | Release date: | 2015-09-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Gaining insight into the catalysis by GH20 lacto-N-biosidase using small molecule inhibitors and structural analysis
Chem.Commun.(Camb.), 51, 2015
|
|
5BXP
 
 | | LNBase in complex with LNB-LOGNAc | | Descriptor: | Lacto-N-biosidase, SULFATE ION, beta-D-galactopyranose-(1-3)-N-acetylglucosaminono-1,5-lactone (Z)-oxime | | Authors: | Ito, T, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-06-09 | | Release date: | 2015-09-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Gaining insight into the catalysis by GH20 lacto-N-biosidase using small molecule inhibitors and structural analysis
Chem.Commun.(Camb.), 51, 2015
|
|
5BXS
 
 | | LNBase in complex with LNB-NHAcCAS | | Descriptor: | 6-ACETAMIDO-6-DEOXY-CASTANOSPERMINE, Lacto-N-biosidase, SULFATE ION, ... | | Authors: | Ito, T, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-06-09 | | Release date: | 2015-09-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Gaining insight into the catalysis by GH20 lacto-N-biosidase using small molecule inhibitors and structural analysis
Chem.Commun.(Camb.), 51, 2015
|
|
5BXR
 
 | | LNBase in complex with LNB-NHAcDNJ | | Descriptor: | 2-ACETAMIDO-1,2-DIDEOXYNOJIRMYCIN, Lacto-N-biosidase, SULFATE ION, ... | | Authors: | Ito, T, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-06-09 | | Release date: | 2015-09-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Gaining insight into the catalysis by GH20 lacto-N-biosidase using small molecule inhibitors and structural analysis
Chem.Commun.(Camb.), 51, 2015
|
|
7CD0
 
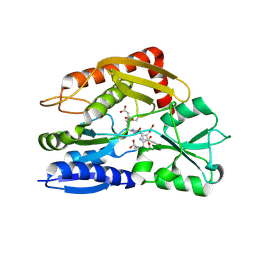 | | Crystal structure of the 2-iodoporphobilinogen-bound ES2 intermediate form of human hydroxymethylbilane synthase | | Descriptor: | 3-[4-(2-hydroxy-2-oxoethyl)-5-[[4-(2-hydroxy-2-oxoethyl)-5-[[4-(2-hydroxy-2-oxoethyl)-5-[[4-(2-hydroxy-2-oxoethyl)-3-(3-hydroxy-3-oxopropyl)-5-methyl-1~{H}-pyrrol-2-yl]methyl]-3-(3-hydroxy-3-oxopropyl)-1~{H}-pyrrol-2-yl]methyl]-3-(3-hydroxy-3-oxopropyl)-1~{H}-pyrrol-2-yl]methyl]-1~{H}-pyrrol-3-yl]propanoic acid, 3-[5-(aminomethyl)-4-(carboxymethyl)-2-iodo-1H-pyrrol-3-yl]propanoic acid, Porphobilinogen deaminase | | Authors: | Sato, H, Sugishima, M, Wada, K, Hirabayashi, K, Tsukaguchi, M. | | Deposit date: | 2020-06-18 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal structures of hydroxymethylbilane synthase complexed with a substrate analog: a single substrate-binding site for four consecutive condensation steps.
Biochem.J., 478, 2021
|
|
7CCZ
 
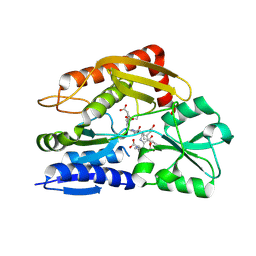 | | Crystal structure of the ES2 intermediate form of human hydroxymethylbilane synthase | | Descriptor: | 3-[4-(2-hydroxy-2-oxoethyl)-5-[[4-(2-hydroxy-2-oxoethyl)-5-[[4-(2-hydroxy-2-oxoethyl)-5-[[4-(2-hydroxy-2-oxoethyl)-3-(3-hydroxy-3-oxopropyl)-5-methyl-1~{H}-pyrrol-2-yl]methyl]-3-(3-hydroxy-3-oxopropyl)-1~{H}-pyrrol-2-yl]methyl]-3-(3-hydroxy-3-oxopropyl)-1~{H}-pyrrol-2-yl]methyl]-1~{H}-pyrrol-3-yl]propanoic acid, Porphobilinogen deaminase | | Authors: | Sato, H, Sugishima, M, Wada, K, Hirabayashi, K, Tsukaguchi, M. | | Deposit date: | 2020-06-18 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structures of hydroxymethylbilane synthase complexed with a substrate analog: a single substrate-binding site for four consecutive condensation steps.
Biochem.J., 478, 2021
|
|
7CCX
 
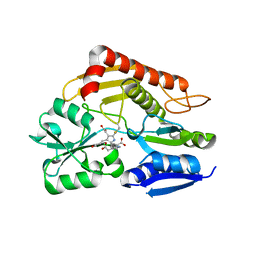 | | Crystal structure of the holo form of human hydroxymethylbilane synthase | | Descriptor: | 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, Porphobilinogen deaminase | | Authors: | Sato, H, Sugishima, M, Wada, K, Hirabayashi, K, Tsukaguchi, M. | | Deposit date: | 2020-06-18 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structures of hydroxymethylbilane synthase complexed with a substrate analog: a single substrate-binding site for four consecutive condensation steps.
Biochem.J., 478, 2021
|
|
7CCY
 
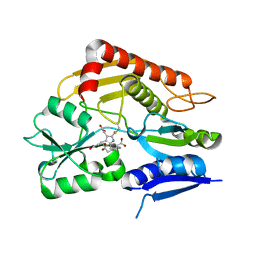 | | Crystal structure of the 2-iodoporphobilinogen-bound holo form of human hydroxymethylbilane synthase | | Descriptor: | 3-[5-(aminomethyl)-4-(carboxymethyl)-2-iodo-1H-pyrrol-3-yl]propanoic acid, 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, Porphobilinogen deaminase | | Authors: | Sato, H, Sugishima, M, Wada, K, Hirabayashi, K, Tsukaguchi, M. | | Deposit date: | 2020-06-18 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of hydroxymethylbilane synthase complexed with a substrate analog: a single substrate-binding site for four consecutive condensation steps.
Biochem.J., 478, 2021
|
|
2Z8F
 
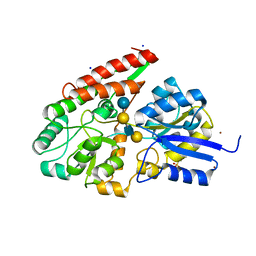 | | The galacto-N-biose-/lacto-N-biose I-binding protein (GL-BP) of the ABC transporter from Bifidobacterium longum in complex with lacto-N-tetraose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Galacto-N-biose/lacto-N-biose I transporter substrate-binding protein, SODIUM ION, ... | | Authors: | Suzuki, R, Wada, J, Katayama, T, Fushinobu, S. | | Deposit date: | 2007-09-05 | | Release date: | 2008-03-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and thermodynamic analyses of solute-binding Protein from Bifidobacterium longum specific for core 1 disaccharide and lacto-N-biose I.
J.Biol.Chem., 283, 2008
|
|
2Z8D
 
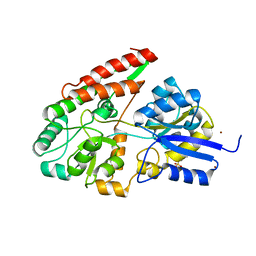 | | The galacto-N-biose-/lacto-N-biose I-binding protein (GL-BP) of the ABC transporter from Bifidobacterium longum in complex with lacto-N-biose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Galacto-N-biose/lacto-N-biose I transporter substrate-binding protein, ZINC ION, ... | | Authors: | Suzuki, R, Wada, J, Katayama, T, Fushinobu, S. | | Deposit date: | 2007-09-05 | | Release date: | 2008-03-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and thermodynamic analyses of solute-binding Protein from Bifidobacterium longum specific for core 1 disaccharide and lacto-N-biose I.
J.Biol.Chem., 283, 2008
|
|
2Z8E
 
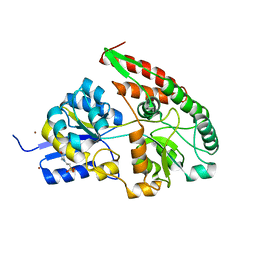 | | The galacto-N-biose-/lacto-N-biose I-binding protein (GL-BP) of the ABC transporter from Bifidobacterium longum in complex with galacto-N-biose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Galacto-N-biose/lacto-N-biose I transporter substrate-binding protein, ZINC ION, ... | | Authors: | Suzuki, R, Wada, J, Katayama, T, Fushinobu, S. | | Deposit date: | 2007-09-05 | | Release date: | 2008-03-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural and thermodynamic analyses of solute-binding Protein from Bifidobacterium longum specific for core 1 disaccharide and lacto-N-biose I.
J.Biol.Chem., 283, 2008
|
|
8HHQ
 
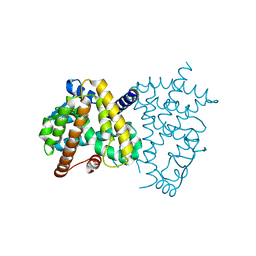 | |
8HHP
 
 | |
2VV3
 
 | | hPPARgamma Ligand binding domain in complex with 4-oxoDHA | | Descriptor: | (6E,10Z,13Z,16Z,19Z)-4-oxodocosa-6,10,13,16,19-pentaenoic acid, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR GAMMA | | Authors: | Itoh, T, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2008-06-02 | | Release date: | 2008-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural Basis for the Activation of Ppargamma by Oxidized Fatty Acids.
Nat.Struct.Mol.Biol., 15, 2008
|
|
2VSR
 
 | | hPPARgamma Ligand binding domain in complex with 9-(S)-HODE | | Descriptor: | (9S,10E,12Z)-9-hydroxyoctadeca-10,12-dienoic acid, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR GAMMA | | Authors: | Itoh, T, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2008-04-29 | | Release date: | 2008-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Basis for the Activation of Pparg by Oxidised Fatty Acids
Nat.Struct.Mol.Biol., 15, 2008
|
|
2VV0
 
 | | hPPARgamma Ligand binding domain in complex with DHA | | Descriptor: | DOCOSA-4,7,10,13,16,19-HEXAENOIC ACID, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR GAMMA | | Authors: | Itoh, T, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2008-06-02 | | Release date: | 2008-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Basis for the Activation of Pparg by Oxidised Fatty Acids
Nat.Struct.Mol.Biol., 15, 2008
|
|
