4IBG
 
 | | Ebola virus VP35 bound to small molecule | | Descriptor: | GLYCEROL, PHOSPHATE ION, Polymerase cofactor VP35, ... | | Authors: | Brown, C.S, Leung, D.W, Xu, W, Borek, D.M, Otwinowski, Z, Ramanan, P, Stubbs, A.J, Peterson, D.S, Binning, J.M, Amarasinghe, G.K. | | Deposit date: | 2012-12-08 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.413 Å) | | Cite: | In Silico Derived Small Molecules Bind the Filovirus VP35 Protein and Inhibit Its Polymerase Cofactor Activity.
J.Mol.Biol., 426, 2014
|
|
4IBD
 
 | | Ebola virus VP35 bound to small molecule | | Descriptor: | 5-[(2R)-3-benzoyl-2-(4-bromothiophen-2-yl)-4-hydroxy-5-oxo-2,5-dihydro-1H-pyrrol-1-yl]-2-methylbenzoic acid, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Brown, C.S, Leung, D.W, Xu, W, Borek, D.M, Otwinowski, Z, Ramanan, P, Stubbs, A.J, Peterson, D.S, Binning, J.M, Amarasinghe, G.K. | | Deposit date: | 2012-12-08 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | In Silico Derived Small Molecules Bind the Filovirus VP35 Protein and Inhibit Its Polymerase Cofactor Activity.
J.Mol.Biol., 426, 2014
|
|
4IBB
 
 | | Ebola virus VP35 bound to small molecule | | Descriptor: | Polymerase cofactor VP35, {4-[(5R)-3-hydroxy-2-oxo-4-(thiophen-2-ylcarbonyl)-5-(2,4,5-trimethylphenyl)-2,5-dihydro-1H-pyrrol-1-yl]phenyl}acetic acid | | Authors: | Brown, C.S, Leung, D.W, Xu, W, Borek, D.M, Otwinowski, Z, Ramanan, P, Stubbs, A.J, Peterson, D.S, Binning, J.M, Amarasinghe, G.K, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-12-08 | | Release date: | 2014-02-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | In Silico Derived Small Molecules Bind the Filovirus VP35 Protein and Inhibit Its Polymerase Cofactor Activity.
J.Mol.Biol., 426, 2014
|
|
4IBI
 
 | | Ebola virus VP35 bound to small molecule | | Descriptor: | 3-{(2S)-2-(7-chloro-1,3-benzodioxol-5-yl)-4-hydroxy-5-oxo-3-[3-(trifluoromethyl)benzoyl]-2,5-dihydro-1H-pyrrol-1-yl}benzoic acid, Polymerase cofactor VP35 | | Authors: | Brown, C.S, Leung, D.W, Xu, W, Borek, D.M, Otwinowski, Z, Ramanan, P, Stubbs, A.J, Peterson, D.S, Binning, J.M, Amarasinghe, G.K. | | Deposit date: | 2012-12-08 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.473 Å) | | Cite: | In Silico Derived Small Molecules Bind the Filovirus VP35 Protein and Inhibit Its Polymerase Cofactor Activity.
J.Mol.Biol., 426, 2014
|
|
4IBF
 
 | | Ebola virus VP35 bound to small molecule | | Descriptor: | (4-{(2R)-2-(4-bromothiophen-2-yl)-3-[(5-chlorothiophen-2-yl)carbonyl]-4-hydroxy-5-oxo-2,5-dihydro-1H-pyrrol-1-yl}phenyl)acetic acid, Polymerase cofactor VP35 | | Authors: | Brown, C.S, Leung, D.W, Xu, W, Borek, D.M, Otwinowski, Z, Ramanan, P, Stubbs, A.J, Peterson, D.S, Binning, J.M, Amarasinghe, G.K, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-12-08 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.291 Å) | | Cite: | In Silico Derived Small Molecules Bind the Filovirus VP35 Protein and Inhibit Its Polymerase Cofactor Activity.
J.Mol.Biol., 426, 2014
|
|
4IBJ
 
 | | Ebola virus VP35 bound to small molecule | | Descriptor: | 3-{(5S)-3-hydroxy-2-oxo-4-[3-(trifluoromethyl)benzoyl]-5-[3-(trifluoromethyl)phenyl]-2,5-dihydro-1H-pyrrol-1-yl}benzoic acid, Polymerase cofactor VP35 | | Authors: | Brown, C.S, Leung, D.W, Xu, W, Borek, D.M, Otwinowski, Z, Ramanan, P, Stubbs, A.J, Peterson, D.S, Binning, J.M, Amarasinghe, G.K. | | Deposit date: | 2012-12-08 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | In Silico Derived Small Molecules Bind the Filovirus VP35 Protein and Inhibit Its Polymerase Cofactor Activity.
J.Mol.Biol., 426, 2014
|
|
4IBE
 
 | | Ebola virus VP35 bound to small molecule | | Descriptor: | 5-[(2R)-3-benzoyl-2-(4-bromothiophen-2-yl)-4-hydroxy-5-oxo-2,5-dihydro-1H-pyrrol-1-yl]-2-chlorobenzoic acid, GLYCEROL, Polymerase cofactor VP35 | | Authors: | Brown, C.S, Leung, D.W, Xu, W, Borek, D.M, Otwinowski, Z, Ramanan, P, Stubbs, A.J, Peterson, D.S, Binning, J.M, Amarasinghe, G.K, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-12-08 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | In Silico Derived Small Molecules Bind the Filovirus VP35 Protein and Inhibit Its Polymerase Cofactor Activity.
J.Mol.Biol., 426, 2014
|
|
4IJF
 
 | | Crystal structure of the Zaire ebolavirus VP35 interferon inhibitory domain K222A/R225A/K248A/K251A mutant | | Descriptor: | Polymerase cofactor VP35 | | Authors: | Binning, J.B, Wang, T, Leung, D.W, Xu, W, Borek, D, Amarasinghe, G.K. | | Deposit date: | 2012-12-21 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.506 Å) | | Cite: | Development of RNA Aptamers Targeting Ebola Virus VP35.
Biochemistry, 52, 2013
|
|
4IBK
 
 | | Ebola virus VP35 bound to small molecule | | Descriptor: | 3-{(2S)-2-(7-chloro-1,3-benzodioxol-5-yl)-3-[(5-chlorothiophen-2-yl)carbonyl]-4-hydroxy-5-oxo-2,5-dihydro-1H-pyrrol-1-yl}benzoic acid, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Brown, C.S, Leung, D.W, Xu, W, Borek, D.M, Otwinowski, Z, Ramanan, P, Stubbs, A.J, Peterson, D.S, Binning, J.M, Amarasinghe, G.K. | | Deposit date: | 2012-12-08 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | In Silico Derived Small Molecules Bind the Filovirus VP35 Protein and Inhibit Its Polymerase Cofactor Activity.
J.Mol.Biol., 426, 2014
|
|
4IJE
 
 | | Crystal structure of the Zaire ebolavirus VP35 interferon inhibitory domain R312A/K319A/R322A mutant | | Descriptor: | PHOSPHATE ION, POTASSIUM ION, Polymerase cofactor VP35, ... | | Authors: | Binning, J.B, Wang, T, Leung, D.W, Xu, W, Borek, D, Amarasinghe, G.K. | | Deposit date: | 2012-12-21 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Development of RNA Aptamers Targeting Ebola Virus VP35.
Biochemistry, 52, 2013
|
|
4IBC
 
 | | Ebola virus VP35 bound to small molecule | | Descriptor: | DIMETHYL SULFOXIDE, Polymerase cofactor VP35, {4-[(2R)-3-(2-chlorobenzoyl)-2-(2-chlorophenyl)-4-hydroxy-5-oxo-2,5-dihydro-1H-pyrrol-1-yl]phenyl}acetic acid | | Authors: | Brown, C.S, Leung, D.W, Xu, W, Borek, D.M, Otwinowski, Z, Ramanan, P, Stubbs, A.J, Peterson, D.S, Binning, J.M, Amarasinghe, G.K. | | Deposit date: | 2012-12-08 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.745 Å) | | Cite: | In Silico Derived Small Molecules Bind the Filovirus VP35 Protein and Inhibit Its Polymerase Cofactor Activity.
J.Mol.Biol., 426, 2014
|
|
4M83
 
 | | Ensemble refinement of protein crystal structure (2IYF) of macrolide glycosyltransferases OleD complexed with UDP and Erythromycin A | | Descriptor: | ERYTHROMYCIN A, MAGNESIUM ION, Oleandomycin glycosyltransferase, ... | | Authors: | Wang, F, Helmich, K.E, Xu, W, Singh, S, Olmos Jr, J.L, Martinez iii, E, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2013-08-12 | | Release date: | 2013-09-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Crystal structure of macrolide glycosyltransferases OleD
To be Published
|
|
4M60
 
 | | Crystal structure of macrolide glycosyltransferases OleD | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Oleandomycin glycosyltransferase, SODIUM ION | | Authors: | Olmos Jr, J.L, Martinez III, E, Wang, F, Helmich, K.E, Singh, S, Xu, W, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2013-08-08 | | Release date: | 2013-09-04 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of macrolide glycosyltransferases OleD
To be Published
|
|
4M7P
 
 | | Ensemble refinement of protein crystal structure of macrolide glycosyltransferases OleD | | Descriptor: | Oleandomycin glycosyltransferase, SODIUM ION | | Authors: | Wang, F, Helmich, K.E, Xu, W, Singh, S, Olmos Jr, J.L, Martinez iii, E, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2013-08-12 | | Release date: | 2013-09-11 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of macrolide glycosyltransferases OleD
To be Published
|
|
4NA4
 
 | | Crystal structure of mouse poly(ADP-ribose) glycohydrolase (PARG) catalytic domain with ADP-HPD | | Descriptor: | 5'-O-[(S)-{[(S)-{[(2R,3R,4S)-3,4-DIHYDROXYPYRROLIDIN-2-YL]METHOXY}(HYDROXY)PHOSPHORYL]OXY}(HYDROXY)PHOSPHORYL]ADENOSINE, IODIDE ION, Poly(ADP-ribose) glycohydrolase | | Authors: | Wang, Z, Cheng, Z, Xu, W. | | Deposit date: | 2013-10-21 | | Release date: | 2014-01-29 | | Last modified: | 2014-09-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic and biochemical analysis of the mouse poly(ADP-ribose) glycohydrolase.
Plos One, 9, 2014
|
|
6KHX
 
 | | Crystal structure of Prx from Akkermansia muciniphila | | Descriptor: | CALCIUM ION, Peroxiredoxin | | Authors: | Li, M, Wang, J, Xu, W, Wang, Y, Zhang, M, Wang, M. | | Deposit date: | 2019-07-16 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Crystal structure of Akkermansia muciniphila peroxiredoxin reveals a novel regulatory mechanism of typical 2-Cys Prxs by a distinct loop.
Febs Lett., 594, 2020
|
|
6LND
 
 | | Crystal structure of transposition protein TniQ | | Descriptor: | ZINC ION, transposition protein TniQ | | Authors: | Wang, B, Xu, W, Yang, H. | | Deposit date: | 2019-12-28 | | Release date: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural basis of a Tn7-like transposase recruitment and DNA loading to CRISPR-Cas surveillance complex.
Cell Res., 30, 2020
|
|
6LNB
 
 | | CryoEM structure of Cascade-TniQ-dsDNA complex | | Descriptor: | CRISPR RNA (60-MER), CRISPR-associated protein Cas6, CRISPR-associated protein Cas7, ... | | Authors: | Wang, B, Xu, W, Yang, H. | | Deposit date: | 2019-12-28 | | Release date: | 2020-02-19 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structural basis of a Tn7-like transposase recruitment and DNA loading to CRISPR-Cas surveillance complex.
Cell Res., 30, 2020
|
|
6LNC
 
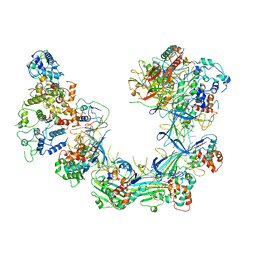 | | CryoEM structure of Cascade-TniQ complex | | Descriptor: | CRISPR RNA (60-MER), CRISPR-associated protein Cas6, CRISPR-associated protein Cas7, ... | | Authors: | Wang, B, Xu, W, Yang, H. | | Deposit date: | 2019-12-28 | | Release date: | 2020-02-19 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structural basis of a Tn7-like transposase recruitment and DNA loading to CRISPR-Cas surveillance complex.
Cell Res., 30, 2020
|
|
7ESH
 
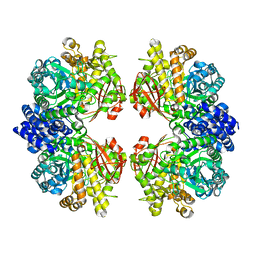 | | Crystal structure of amylosucrase from Calidithermus timidus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, amylosucrase | | Authors: | Tian, Y, Hou, X, Ni, D, Xu, W, Guang, C, Zhang, W, Rao, Y, Mu, W. | | Deposit date: | 2021-05-10 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structure-based interface engineering methodology in designing a thermostable amylose-forming transglucosylase
J.Biol.Chem., 298, 2022
|
|
5WZZ
 
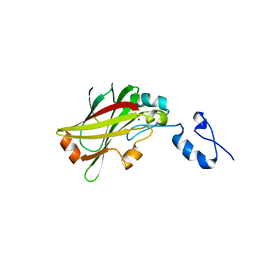 | | The SIAH E3 ubiquitin ligases promote Wnt/ beta-catenin signaling through mediating Wnt-induced Axin degradation | | Descriptor: | Axin-1, E3 ubiquitin-protein ligase SIAH1, ZINC ION | | Authors: | Ji, L, Jiang, B, Jiang, X, Charlat, O, Chen, A, Mickanin, C, Bauer, A, Xu, W, Yan, X.-X, Cong, F. | | Deposit date: | 2017-01-19 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | The SIAH E3 ubiquitin ligases promote Wnt/ beta-catenin signaling through mediating Wnt-induced Axin degradation
Genes Dev., 31, 2017
|
|
6BMG
 
 | | Structure of Recombinant Dwarf Sperm Whale Myoglobin (Oxy) | | Descriptor: | ACETATE ION, CADMIUM ION, Myoglobin, ... | | Authors: | Samuel, P.P, Miller, M.D, Xu, W, Alvarado, S, Phillips Jr, G.N, Olson, J.S. | | Deposit date: | 2017-11-14 | | Release date: | 2017-11-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structure of Recombinant Dwarf Sperm Whale Myoglobin (Oxy)
To Be Published
|
|
6BUI
 
 | |
6C6K
 
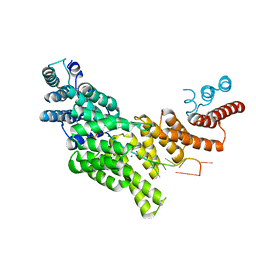 | | Structural basis for preferential recognition of cap 0 RNA by a human IFIT1-IFIT3 protein complex | | Descriptor: | Interferon-induced protein with tetratricopeptide repeats 1, Interferon-induced protein with tetratricopeptide repeats 3, MAGNESIUM ION, ... | | Authors: | Amarasinghe, G.K, Leung, D.W, Johnson, B, Xu, W. | | Deposit date: | 2018-01-18 | | Release date: | 2018-04-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Human IFIT3 Modulates IFIT1 RNA Binding Specificity and Protein Stability.
Immunity, 48, 2018
|
|
6BUG
 
 | |
