5UTR
 
 | | Crystal structure of Burkholderia cenocepacia family 3 glycoside hydrolase (NagZ) bound to (3S,4R,5R,6S)-3-butyryl-4,5,6-trihydroxyazepane | | Descriptor: | Beta-hexosaminidase, N-[(3S,4R,5R,6S)-4,5,6-trihydroxyazepan-3-yl]butanamide | | Authors: | Vadlamani, G, Mark, B.L. | | Deposit date: | 2017-02-15 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Conformational flexibility of the glycosidase NagZ allows it to bind structurally diverse inhibitors to suppress beta-lactam antibiotic resistance.
Protein Sci., 26, 2017
|
|
7QQF
 
 | | Crystal structure of unliganded MYORG | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Meek, R.W, Davies, G.J. | | Deposit date: | 2022-01-07 | | Release date: | 2022-08-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | The primary familial brain calcification-associated protein MYORG is an alpha-galactosidase with restricted substrate specificity.
Plos Biol., 20, 2022
|
|
7QQH
 
 | | Crystal structure of MYORG (D520N) in complex with Gal-a1,4-Glc | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Meek, R.W, Davies, G.J. | | Deposit date: | 2022-01-07 | | Release date: | 2022-08-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The primary familial brain calcification-associated protein MYORG is an alpha-galactosidase with restricted substrate specificity.
Plos Biol., 20, 2022
|
|
7QQG
 
 | | Crystal structure of MYORG bound to 1-deoxygalactonojirimycin | | Descriptor: | (2R,3S,4R,5S)-2-(hydroxymethyl)piperidine-3,4,5-triol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Meek, R.W, Davies, G.J. | | Deposit date: | 2022-01-07 | | Release date: | 2022-08-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | The primary familial brain calcification-associated protein MYORG is an alpha-galactosidase with restricted substrate specificity.
Plos Biol., 20, 2022
|
|
8AWK
 
 | | Structure of recombinant human beta-glucocerebrosidase in complex with D-carbaxylosyl chloride | | Descriptor: | (2~{S},3~{S},4~{R})-cyclohex-5-ene-1,2,3,4-tetrol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rowland, R.J, Davies, G.J. | | Deposit date: | 2022-08-30 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Single turnover covalent inhibitors for functional chaperoning of lysosomal glycoside hydrolases
To be published
|
|
8AWR
 
 | | Structure of recombinant human beta-glucocerebrosidase in complex with L-carbaxylosyl chloride | | Descriptor: | (1~{S},2~{R},3~{S},6~{S})-6-chloranylcyclohex-4-ene-1,2,3-triol, (1~{S},2~{S},3~{S},4~{R})-cyclohexane-1,2,3,4-tetrol, 1,2-ETHANEDIOL, ... | | Authors: | Rowland, R.J, Davies, G.J. | | Deposit date: | 2022-08-30 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Single turnover covalent inhibitors for functional chaperoning of lysosomal glycoside hydrolases
To be published
|
|
8AX3
 
 | | Structure of recombinant human beta-glucocerebrosidase in complex with L-carbaxylosyl fluoride | | Descriptor: | (1~{S},2~{R},3~{S},6~{S})-6-fluoranylcyclohex-4-ene-1,2,3-triol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rowland, R.J, Davies, G.J. | | Deposit date: | 2022-08-30 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Single turnover covalent inhibitors for functional chaperoning of lysosomal glycoside hydrolases
To be published
|
|
8CM9
 
 | | Structure of human O-GlcNAc transferase in complex with UDP and tP11 | | Descriptor: | PHE-MET-PRO-LYS-TYR-SER-ILE, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit, URIDINE-5'-DIPHOSPHATE | | Authors: | Meek, R.W, Davies, G.J. | | Deposit date: | 2023-02-18 | | Release date: | 2023-10-04 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Phage display uncovers a sequence motif that drives polypeptide binding to a conserved regulatory exosite of O-GlcNAc transferase.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
3GS6
 
 | | Vibrio Cholerea family 3 glycoside hydrolase (NagZ)in complex with N-butyryl-PUGNAc | | Descriptor: | Beta-hexosaminidase, GLYCEROL, [[(3R,4R,5S,6R)-3-(BUTANOYLAMINO)-4,5-DIHYDROXY-6-(HYDROXYMETHYL)OXAN-2-YLIDENE]AMINO] N-PHENYLCARBAMATE | | Authors: | Balcewich, M.D, Mark, B.L. | | Deposit date: | 2009-03-26 | | Release date: | 2009-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insight into a strategy for attenuating AmpC-mediated beta-lactam resistance: structural basis for selective inhibition of the glycoside hydrolase NagZ.
Protein Sci., 18, 2009
|
|
3GSM
 
 | | Vibrio cholerae family 3 glycoside hydrolase (NagZ) bound to N-Valeryl-PUGNAc | | Descriptor: | Beta-hexosaminidase, GLYCEROL, [[(3R,4R,5S,6R)-4,5-dihydroxy-6-(hydroxymethyl)-3-(pentanoylamino)oxan-2-ylidene]amino] N-phenylcarbamate | | Authors: | Balcewich, M.D, Mark, B.L. | | Deposit date: | 2009-03-27 | | Release date: | 2009-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Insight into a strategy for attenuating AmpC-mediated beta-lactam resistance: structural basis for selective inhibition of the glycoside hydrolase NagZ.
Protein Sci., 18, 2009
|
|
3KOT
 
 | |
3KOS
 
 | |
3QBW
 
 | | Crystal structure of pseudomonas aeruginosa 1,6-anhydro-n-actetylmuramic acid kinase (ANMK) bound to adenosine diphosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Anhydro-N-acetylmuramic acid kinase, SULFATE ION | | Authors: | Bacik, J.P, Martin, D.R, Mark, B.L. | | Deposit date: | 2011-01-14 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Molecular Basis of 1,6-Anhydro Bond Cleavage and Phosphoryl Transfer by Pseudomonas aeruginosa 1,6-Anhydro-N-acetylmuramic Acid Kinase.
J.Biol.Chem., 286, 2011
|
|
3QBX
 
 | | Crystal structure of pseudomonas aeruginosa 1,6-anhydro-n-actetylmuramic acid kinase (ANMK) bound to 1,6-anhydro-n-actetylmuramic acid | | Descriptor: | 2-(2-ACETYLAMINO-4-HYDROXY-6,8-DIOXA-BICYCLO[3.2.1]OCT-3-YLOXY)-PROPIONIC ACID, Anhydro-N-acetylmuramic acid kinase, SULFATE ION | | Authors: | Bacik, J.P, Martin, D.R, Mark, B.L. | | Deposit date: | 2011-01-14 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular Basis of 1,6-Anhydro Bond Cleavage and Phosphoryl Transfer by Pseudomonas aeruginosa 1,6-Anhydro-N-acetylmuramic Acid Kinase.
J.Biol.Chem., 286, 2011
|
|
2VSN
 
 | | Structure and topological arrangement of an O-GlcNAc transferase homolog: insight into molecular control of intracellular glycosylation | | Descriptor: | URIDINE-5'-DIPHOSPHATE, XCOGT | | Authors: | Martinez-Fleites, C, Macauley, M.S, He, Y, Shen, D, Vocadlo, D, Davies, G.J. | | Deposit date: | 2008-04-28 | | Release date: | 2008-06-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of an O-Glcnac Transferase Homolog Provides Insight Into Intracellular Glycosylation.
Nat.Struct.Mol.Biol., 15, 2008
|
|
4MSS
 
 | | Crystal structure of Burkholderia cenocepacia family 3 glycoside hydrolase (NagZ) bound to (3S,4R,5R,6S)-3-acetamido-4,5,6-trihydroxyazepane | | Descriptor: | Beta-hexosaminidase 1, GLYCEROL, N-[(3S,4R,5R,6S)-4,5,6-trihydroxyazepan-3-yl]acetamide | | Authors: | Vadlamani, G, Mark, B.L. | | Deposit date: | 2013-09-18 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Selective trihydroxyazepane NagZ inhibitors increase sensitivity of Pseudomonas aeruginosa to beta-lactams.
Chem.Commun.(Camb.), 49, 2013
|
|
4MO5
 
 | | Crystal structure of AnmK bound to AMPPCP and anhMurNAc | | Descriptor: | 2-(2-ACETYLAMINO-4-HYDROXY-6,8-DIOXA-BICYCLO[3.2.1]OCT-3-YLOXY)-PROPIONIC ACID, Anhydro-N-acetylmuramic acid kinase, MAGNESIUM ION, ... | | Authors: | Bacik, J.P, Mark, B.L. | | Deposit date: | 2013-09-11 | | Release date: | 2014-01-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Conformational Itinerary of Pseudomonas aeruginosa 1,6-Anhydro-N-acetylmuramic Acid Kinase during Its Catalytic Cycle.
J.Biol.Chem., 289, 2014
|
|
4N39
 
 | |
4MO4
 
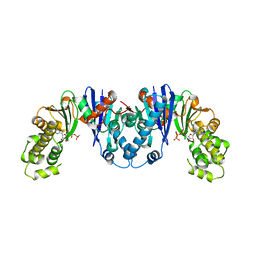 | | Crystal structure of AnmK bound to AMPPCP | | Descriptor: | Anhydro-N-acetylmuramic acid kinase, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Bacik, J.P, Mark, B.L. | | Deposit date: | 2013-09-11 | | Release date: | 2014-01-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Conformational Itinerary of Pseudomonas aeruginosa 1,6-Anhydro-N-acetylmuramic Acid Kinase during Its Catalytic Cycle.
J.Biol.Chem., 289, 2014
|
|
4N3B
 
 | | Crystal Structure of human O-GlcNAc Transferase bound to a peptide from HCF-1 pro-repeat2(1-26)E10Q and UDP-5SGlcNAc | | Descriptor: | (2S,3R,4R,5S,6R)-3-(acetylamino)-4,5-dihydroxy-6-(hydroxymethyl)tetrahydro-2H-thiopyran-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate, Host cell factor 1, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit | | Authors: | Lazarus, M.B, Herr, W, Walker, S. | | Deposit date: | 2013-10-06 | | Release date: | 2014-01-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | HCF-1 is cleaved in the active site of O-GlcNAc transferase.
Science, 342, 2013
|
|
4N3A
 
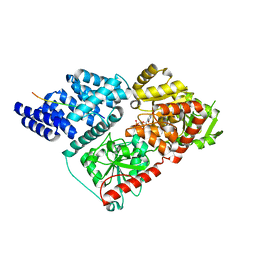 | |
4N3C
 
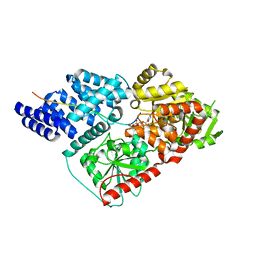 | |
4OJN
 
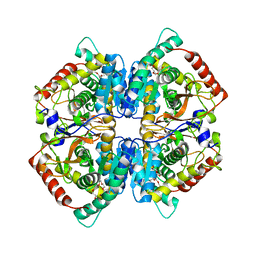 | | Crystal structure of human muscle L-lactate dehydrogenase | | Descriptor: | GLYCEROL, L-lactate dehydrogenase A chain, PENTAETHYLENE GLYCOL | | Authors: | Kolappan, S, Craig, L. | | Deposit date: | 2014-01-21 | | Release date: | 2014-12-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of lactate dehydrogenase A (LDHA) in apo, ternary and inhibitor-bound forms.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4OKN
 
 | | Crystal structure of human muscle L-lactate dehydrogenase, ternary complex with NADH and oxalate | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, KANAMYCIN A, L-lactate dehydrogenase A chain, ... | | Authors: | Kolappan, S, Craig, L. | | Deposit date: | 2014-01-22 | | Release date: | 2014-12-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of lactate dehydrogenase A (LDHA) in apo, ternary and inhibitor-bound forms.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4AIS
 
 | | A complex structure of BtGH84 | | Descriptor: | GLYCEROL, GLYCOLIC ACID, O-GLCNACASE BT_4395 | | Authors: | He, Y, Davies, G.J. | | Deposit date: | 2012-02-13 | | Release date: | 2012-06-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Metabolism of Vertebrate Amino Sugars with N-Glycolyl Groups: Intracellular Beta-O-Linked N-Glycolylglucosamine (Glcngc), Udp-Glcngc, and the Biochemical and Structural Rationale for the Substrate Tolerance of Beta-O-Linked Beta-N-Acetylglucosaminidase.
J.Biol.Chem., 287, 2012
|
|
