4QG8
 
 | | crystal structure of PKM2-K305Q mutant | | Descriptor: | GLYCEROL, MAGNESIUM ION, MALONATE ION, ... | | Authors: | Wang, P, Sun, C, Zhu, T, Xu, Y. | | Deposit date: | 2014-05-22 | | Release date: | 2015-02-25 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insight into mechanisms for dynamic regulation of PKM2.
Protein Cell, 6, 2015
|
|
4QG9
 
 | | crystal structure of PKM2-R399E mutant | | Descriptor: | ACETATE ION, MAGNESIUM ION, Pyruvate kinase PKM | | Authors: | Wang, P, Sun, C, Zhu, T, Xu, Y. | | Deposit date: | 2014-05-22 | | Release date: | 2015-02-25 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.381 Å) | | Cite: | Structural insight into mechanisms for dynamic regulation of PKM2.
Protein Cell, 6, 2015
|
|
4RPP
 
 | | crystal structure of PKM2-K422R mutant bound with FBP | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, Pyruvate kinase PKM | | Authors: | Wang, P, Sun, C, Zhu, T, Xu, Y. | | Deposit date: | 2014-10-31 | | Release date: | 2015-02-25 | | Last modified: | 2023-04-05 | | Method: | X-RAY DIFFRACTION (2.585 Å) | | Cite: | Structural insight into mechanisms for dynamic regulation of PKM2.
Protein Cell, 6, 2015
|
|
1TFT
 
 | | NMR Structure of an Antagonists of the XIAP-Caspase-9 Interaction Complexed to the BIR3 domain of XIAP | | Descriptor: | 1-[3,3-DIMETHYL-2-(2-METHYLAMINO-PROPIONYLAMINO)-BUTYRYL]-4-PHENOXY-PYRROLIDINE-2-CARBOXYLIC ACID(1,2,3,4-TETRAHYDRO-NAPHTHALEN-1-YL)-AMIDE, Baculoviral IAP repeat-containing protein 4, ZINC ION | | Authors: | Oost, T.K, Sun, C, Armstrong, R.C, Al-Assaad, A.S, Betz, S.F, Deckwerth, T.L, Elmore, S.W, Meadows, R.P, Olejniczak, E.T, Oleksijew, A.K, Oltersdorf, T, Rosenberg, S.H, Shoemaker, A.R, Zou, H, Fesik, S.W. | | Deposit date: | 2004-05-27 | | Release date: | 2005-05-03 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Discovery of potent antagonists of the antiapoptotic protein XIAP for the treatment of cancer.
J.Med.Chem., 47, 2004
|
|
1TFQ
 
 | | NMR Structure of an Antagonists of the XIAP-Caspase-9 Interaction Complexed to the BIR3 domain of XIAP | | Descriptor: | Baculoviral IAP repeat-containing protein 4, N-METHYLALANYL-3-METHYLVALYL-N-(1,2,3,4-TETRAHYDRONAPHTHALEN-1-YL)PROLINAMIDE, ZINC ION | | Authors: | Oost, T.K, Sun, C, Armstrong, R.C, Al-Assaad, A.S, Betz, S.F, Deckwerth, T.L, Elmore, S.W, Meadows, R.P, Olejniczak, E.T, Oleksijew, A, Oltersdorf, T, Rosenberg, S.H, Shoemaker, A.R, Zou, H, Fesik, S.W. | | Deposit date: | 2004-05-27 | | Release date: | 2004-09-07 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Discovery of Potent Antagonists of the Antiapoptotic Protein XIAP for the Treatment of Cancer.
J.Med.Chem., 47, 2004
|
|
2A9H
 
 | | NMR structural studies of a potassium channel / charybdotoxin complex | | Descriptor: | Voltage-gated potassium channel, charybdotoxin | | Authors: | Yu, L, Sun, C, Song, D, Shen, J, Xu, N, Gunasekera, A, Hajduk, P.J, Olejniczak, E.T. | | Deposit date: | 2005-07-11 | | Release date: | 2006-01-10 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structural studies of a potassium channel-charybdotoxin complex.
Biochemistry, 44, 2005
|
|
2E6W
 
 | | Solution structure and calcium binding properties of EF-hands 3 and 4 of calsenilin | | Descriptor: | CALCIUM ION, Calsenilin | | Authors: | Yu, L, Sun, C, Mendoza, R, Hebert, E, Pereda-Lopez, A, Hajduk, P.J, Olejniczak, E.T. | | Deposit date: | 2007-01-05 | | Release date: | 2007-11-27 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure and calcium-binding properties of EF-hands 3 and 4 of calsenilin.
Protein Sci., 16, 2007
|
|
8J2G
 
 | | Human ZnT1-D47N/D255N mutant | | Descriptor: | Proton-coupled zinc antiporter SLC30A1 | | Authors: | Sun, L, Liu, X, Sun, C, He, B. | | Deposit date: | 2023-04-14 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the human ZnT1-D47N/D255N double mutant
To Be Published
|
|
3NEW
 
 | | p38-alpha complexed with Compound 10 | | Descriptor: | 4-(trifluoromethyl)-3-[3-(trifluoromethyl)phenyl]-1,7-dihydro-6H-pyrazolo[3,4-b]pyridin-6-one, Mitogen-activated protein kinase 14 | | Authors: | Goedken, E.R, Comess, K.M, Sun, C, Argiriadi, M, Jia, Y, Quinn, C.M, Banach, D.L, Marcotte, D, Borhani, D. | | Deposit date: | 2010-06-09 | | Release date: | 2010-12-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Discovery and Characterization of Non-ATP Site Inhibitors of the Mitogen Activated Protein (MAP) Kinases.
Acs Chem.Biol., 6, 2011
|
|
6MX4
 
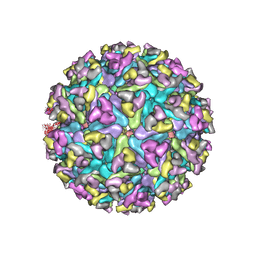 | | CryoEM structure of chimeric Eastern Equine Encephalitis Virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Capsid, ... | | Authors: | Hasan, S.S, Sun, C, Kim, A.S, Watanabe, Y, Chen, C.L, Klose, T, Buda, G, Crispin, M, Diamond, M.S, Klimstra, W.B, Rossmann, M.G. | | Deposit date: | 2018-10-30 | | Release date: | 2018-12-19 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM Structures of Eastern Equine Encephalitis Virus Reveal Mechanisms of Virus Disassembly and Antibody Neutralization.
Cell Rep, 25, 2018
|
|
6MWC
 
 | | CryoEM structure of chimeric Eastern Equine Encephalitis Virus with Fab of EEEV-5 antibody | | Descriptor: | E1, E2, EEEV-5 antibody heavy chain, ... | | Authors: | Hasan, S.S, Sun, C, Kim, A.S, Watanabe, Y, Chen, C.L, Klose, T, Buda, G, Crispin, M, Diamond, M.S, Klimstra, W.B, Rossmann, M.G. | | Deposit date: | 2018-10-29 | | Release date: | 2018-12-19 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Cryo-EM Structures of Eastern Equine Encephalitis Virus Reveal Mechanisms of Virus Disassembly and Antibody Neutralization.
Cell Rep, 25, 2018
|
|
2NW4
 
 | | Crystal Structure of the Rat Androgen Receptor Ligand Binding Domain Complex with BMS-564929 | | Descriptor: | 2-CHLORO-4-[(7R,7AS)-7-HYDROXY-1,3-DIOXOTETRAHYDRO-1H-PYRROLO[1,2-C]IMIDAZOL-2(3H)-YL]-3-METHYLBENZONITRILE, Androgen receptor | | Authors: | Ostrowski, J, Kuhns, J.E, Lupisella, J.A, Manfredi, M.C, Beehler, B.C, Krystek, S.R, Bi, Y, Sun, C, Seethala, R, Golla, R, Sleph, P.G, Fura, A, An, Y, Kish, K.F, Sack, J.S, Mookhtiar, K.A, Grover, G.J, Hamann, L.G. | | Deposit date: | 2006-11-14 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Pharmacological and x-ray structural characterization of a novel selective androgen receptor modulator: potent hyperanabolic stimulation of skeletal muscle with hypostimulation of prostate in rats.
Endocrinology, 148, 2007
|
|
5CPR
 
 | | The novel SUV4-20 inhibitor A-196 verifies a role for epigenetics in genomic integrity | | Descriptor: | 6,7-dichloro-N-cyclopentyl-4-(pyridin-4-yl)phthalazin-1-amine, Histone-lysine N-methyltransferase SUV420H1, S-ADENOSYLMETHIONINE, ... | | Authors: | Jakob, C.G, Upadhyay, A.K, Sun, C. | | Deposit date: | 2015-07-21 | | Release date: | 2017-01-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | The SUV4-20 inhibitor A-196 verifies a role for epigenetics in genomic integrity.
Nat. Chem. Biol., 13, 2017
|
|
1I3O
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX OF XIAP-BIR2 AND CASPASE 3 | | Descriptor: | BACULOVIRAL IAP REPEAT-CONTAINING PROTEIN 4, CASPASE 3, ZINC ION | | Authors: | Riedl, S.J, Renatus, M, Schwarzenbacher, R, Zhou, Q, Sun, C, Fesik, S.W, Liddington, R.C, Salvesen, G.S. | | Deposit date: | 2001-02-15 | | Release date: | 2001-03-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the inhibition of caspase-3 by XIAP.
Cell(Cambridge,Mass.), 104, 2001
|
|
4OQ6
 
 | | Crystal Structure of Human MCL-1 Bound to Inhibitor 4-hydroxy-4'-propylbiphenyl-3-carboxylic acid | | Descriptor: | 4-hydroxy-4'-propylbiphenyl-3-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Petros, A.M, Swann, S.L, Song, D, Swinger, K, Park, C, Zhang, H, Wendt, M.D, Kunzer, A.R, Souers, A.J, Sun, C. | | Deposit date: | 2014-02-07 | | Release date: | 2014-03-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Fragment-based discovery of potent inhibitors of the anti-apoptotic MCL-1 protein.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4OQ5
 
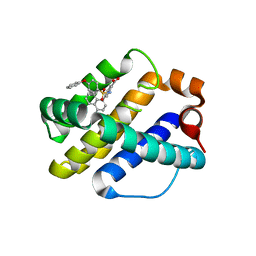 | | Crystal Structure of Human MCL-1 Bound to Inhibitor 4-(4-methylnaphthalen-1-yl)-2-{[(4-phenoxyphenyl)sulfonyl]amino}benzoic acid | | Descriptor: | 4-(4-methylnaphthalen-1-yl)-2-{[(4-phenoxyphenyl)sulfonyl]amino}benzoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Petros, A.M, Swann, S.L, Song, D, Swinger, K, Park, C, Zhang, H, Wendt, M.D, Kunzer, A.R, Souers, A.J, Sun, C. | | Deposit date: | 2014-02-07 | | Release date: | 2014-03-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Fragment-based discovery of potent inhibitors of the anti-apoptotic MCL-1 protein.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
5C56
 
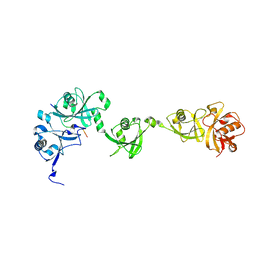 | | Crystal structure of USP7/HAUSP in complex with ICP0 | | Descriptor: | Ubiquitin E3 ligase ICP0, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Cheng, J, Li, Z, Gong, R, Fang, J, Yang, Y, Sun, C, Yang, H, Xu, Y. | | Deposit date: | 2015-06-19 | | Release date: | 2015-07-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.685 Å) | | Cite: | Molecular mechanism for the substrate recognition of USP7.
Protein Cell, 6, 2015
|
|
5V33
 
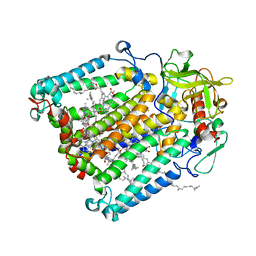 | | R. sphaeroides photosythetic reaction center mutant - Residue L223, Ser to Trp - Room Temperature Structure Solved on X-ray Transparent Microfluidic Chip | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (III) ION, ... | | Authors: | Schieferstein, J.M, Pawate, A.S, Sun, C, Wan, F, Broecker, J, Ernst, O.P, Gennis, R.B, Kenis, P.J.A. | | Deposit date: | 2017-03-06 | | Release date: | 2017-04-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.487 Å) | | Cite: | X-ray transparent microfluidic chips for high-throughput screening and optimization of in meso membrane protein crystallization.
Biomicrofluidics, 11, 2017
|
|
4MGJ
 
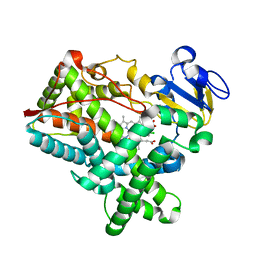 | | Crystal structure of cytochrome P450 2B4 F429H in complex with 4-CPI | | Descriptor: | 4-(4-CHLOROPHENYL)IMIDAZOLE, Cytochrome P450 2B4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yang, Y, Zhang, H, Usharani, D, Bu, W, Im, S, Tarasev, M, Rwere, F, Meagher, J, Sun, C, Stuckey, J, Shaik, S, Waskell, L. | | Deposit date: | 2013-08-28 | | Release date: | 2014-08-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structural and Functional Characterization of a Cytochrome P450 2B4 F429H Mutant with an Axial Thiolate-Histidine Hydrogen Bond.
Biochemistry, 53, 2014
|
|
4QGC
 
 | | crystal structure of PKM2-K422R mutant | | Descriptor: | GLYCEROL, POTASSIUM ION, Pyruvate kinase PKM, ... | | Authors: | Wang, P, Sun, C, Zhu, T, Xu, Y. | | Deposit date: | 2014-05-22 | | Release date: | 2015-02-25 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.296 Å) | | Cite: | Structural insight into mechanisms for dynamic regulation of PKM2.
Protein Cell, 6, 2015
|
|
7K5W
 
 | | Cryo-EM structure of heterologous protein complex loaded Thermotoga maritima encapsulin capsid | | Descriptor: | Maritimacin | | Authors: | Xiong, X, Sun, C, Vago, F.S, Klose, T, Zhu, J, Jiang, W. | | Deposit date: | 2020-09-17 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Cryo-EM Structure of Heterologous Protein Complex Loaded Thermotoga Maritima Encapsulin Capsid.
Biomolecules, 10, 2020
|
|
6MW9
 
 | | CryoEM structure of chimeric Eastern Equine Encephalitis Virus with Fab of EEEV-3 antibody | | Descriptor: | E1, E2, EEEV-3 antibody heavy chain, ... | | Authors: | Hasan, S.S, Sun, C, Kim, A.S, Watanabe, Y, Chen, C.L, Klose, T, Buda, G, Crispin, M, Diamond, M.S, Klimstra, W.B, Rossmann, M.G. | | Deposit date: | 2018-10-29 | | Release date: | 2018-12-19 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Cryo-EM Structures of Eastern Equine Encephalitis Virus Reveal Mechanisms of Virus Disassembly and Antibody Neutralization.
Cell Rep, 25, 2018
|
|
6MWX
 
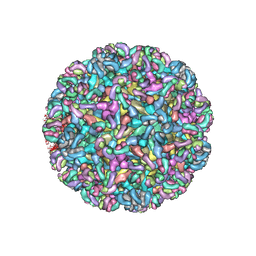 | | CryoEM structure of Chimeric Eastern Equine Encephalitis Virus with Fab of EEEV-69 Antibody | | Descriptor: | E1, E2, EEEV-69 antibody heavy chain, ... | | Authors: | Hasan, S.S, Sun, C, Kim, A.S, Watanabe, Y, Chen, C.L, Klose, T, Buda, G, Crispin, M, Diamond, M.S, Klimstra, W.B, Rossmann, M.G. | | Deposit date: | 2018-10-30 | | Release date: | 2018-12-19 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | Cryo-EM Structures of Eastern Equine Encephalitis Virus Reveal Mechanisms of Virus Disassembly and Antibody Neutralization.
Cell Rep, 25, 2018
|
|
6MUI
 
 | | CryoEM structure of chimeric Eastern Equine Encephalitis Virus with Fab of EEEV-42 antibody | | Descriptor: | E1, E2, EEEV-42 antibody heavy chain, ... | | Authors: | Hasan, S.S, Sun, C, Kim, A.S, Watanabe, Y, Chen, C.L, Klose, T, Buda, G, Crispin, M, Diamond, M.S, Klimstra, W.B, Rossmann, M.G. | | Deposit date: | 2018-10-23 | | Release date: | 2018-12-19 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Cryo-EM Structures of Eastern Equine Encephalitis Virus Reveal Mechanisms of Virus Disassembly and Antibody Neutralization.
Cell Rep, 25, 2018
|
|
6MX7
 
 | | CryoEM structure of chimeric Eastern Equine Encephalitis Virus: Genome-Binding Capsid N-terminal Domain | | Descriptor: | Capsid | | Authors: | Hasan, S.S, Sun, C, Kim, A.S, Watanabe, Y, Chen, C.L, Klose, T, Buda, G, Crispin, M, Diamond, M.S, Klimstra, W.B, Rossmann, M.G. | | Deposit date: | 2018-10-30 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Cryo-EM Structures of Eastern Equine Encephalitis Virus Reveal Mechanisms of Virus Disassembly and Antibody Neutralization.
Cell Rep, 25, 2018
|
|
