1UHV
 
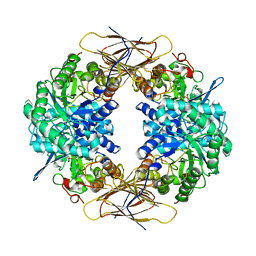 | | Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase | | 分子名称: | 1,5-anhydro-2-deoxy-2-fluoro-D-xylitol, Beta-xylosidase | | 著者 | Yang, J.K, Yoon, H.J, Ahn, H.J, Il Lee, B, Pedelacq, J.D, Liong, E.C, Berendzen, J, Laivenieks, M, Vieille, C, Zeikus, G.J, Vocadlo, D.J, Withers, S.G, Suh, S.W. | | 登録日 | 2003-07-11 | | 公開日 | 2003-12-23 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase.
J.Mol.Biol., 335, 2004
|
|
1TAQ
 
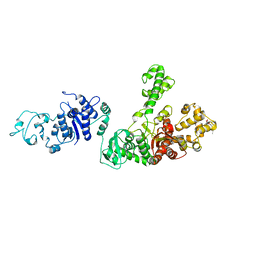 | | STRUCTURE OF TAQ DNA POLYMERASE | | 分子名称: | 2-O-octyl-beta-D-glucopyranose, TAQ DNA POLYMERASE, ZINC ION | | 著者 | Kim, Y, Eom, S.H, Wang, J, Lee, D.-S, Suh, S.W, Steitz, T.A. | | 登録日 | 1996-06-04 | | 公開日 | 1996-12-07 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of Thermus aquaticus DNA polymerase.
Nature, 376, 1995
|
|
1UM0
 
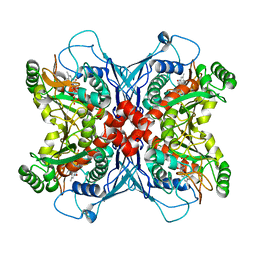 | | Crystal structure of chorismate synthase complexed with FMN | | 分子名称: | Chorismate synthase, FLAVIN MONONUCLEOTIDE | | 著者 | Ahn, H.J, Yoon, H.J, Lee, B, Suh, S.W. | | 登録日 | 2003-09-18 | | 公開日 | 2004-06-01 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal structure of chorismate synthase: a novel FMN-binding protein fold and functional insights
J.Mol.Biol., 336, 2004
|
|
1VC1
 
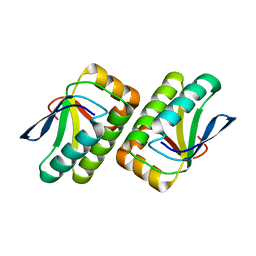 | | Crystal structure of the TM1442 protein from Thermotoga maritima, a homolog of the Bacillus subtilis general stress response anti-anti-sigma factor RsbV | | 分子名称: | Putative anti-sigma factor antagonist TM1442 | | 著者 | Lee, J.Y, Ahn, H.J, Ha, K.S, Suh, S.W. | | 登録日 | 2004-03-03 | | 公開日 | 2004-09-28 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of the TM1442 protein from Thermotoga maritima, a homolog of the Bacillus subtilis general stress response anti-anti-sigma factor RsbV
Proteins, 56, 2004
|
|
1V9P
 
 | | Crystal Structure Of Nad+-Dependent DNA Ligase | | 分子名称: | ADENOSINE MONOPHOSPHATE, DNA ligase, ZINC ION | | 著者 | Lee, J.Y, Chang, C, Song, H.K, Moon, J, Yang, J.K, Kim, H.K, Kwon, S.K, Suh, S.W. | | 登録日 | 2004-01-27 | | 公開日 | 2004-03-30 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Crystal structure of NAD(+)-dependent DNA ligase: modular architecture and functional implications.
Embo J., 19, 2000
|
|
1UMF
 
 | |
4RUB
 
 | | A CRYSTAL FORM OF RIBULOSE-1,5-BISPHOSPHATE CARBOXYLASE(SLASH)OXYGENASE FROM NICOTIANA TABACUM IN THE ACTIVATED STATE | | 分子名称: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, FORMIC ACID, MAGNESIUM ION, ... | | 著者 | Schreuder, H, Cascio, D, Curmi, P.M.G, Eisenberg, D. | | 登録日 | 1990-05-25 | | 公開日 | 1992-10-15 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | A crystal form of ribulose-1,5-bisphosphate carboxylase/oxygenase from Nicotiana tabacum in the activated state.
J.Mol.Biol., 197, 1987
|
|
5ZHF
 
 | | Structure of VanYB unbound | | 分子名称: | D-alanyl-D-alanine carboxypeptidase, GLYCEROL, TETRAETHYLENE GLYCOL, ... | | 著者 | Kim, H.S, Hahn, H. | | 登録日 | 2018-03-13 | | 公開日 | 2018-09-05 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structural basis for the substrate recognition of peptidoglycan pentapeptides by Enterococcus faecalis VanYB.
Int. J. Biol. Macromol., 119, 2018
|
|
1L5X
 
 | | The 2.0-Angstrom resolution crystal structure of a survival protein E (SurE) homolog from Pyrobaculum aerophilum | | 分子名称: | ACETIC ACID, GLYCEROL, Survival protein E | | 著者 | Mura, C, Katz, J.E, Clarke, S.G, Eisenberg, D. | | 登録日 | 2002-03-08 | | 公開日 | 2003-02-25 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure and Function of an Archaeal Homolog of Survival
Protein E (SurE-alpha): An Acid Phosphatase with Purine
Nucleotide Specificity
J.Mol.Biol., 326, 2003
|
|
3JUJ
 
 | |
5YCN
 
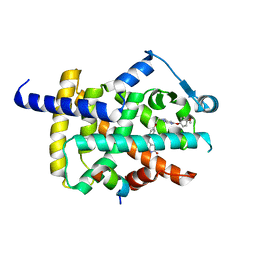 | | Human PPARgamma ligand binding domain complexed with Lobeglitazone | | 分子名称: | (5S)-5-[[4-[2-[[6-(4-methoxyphenoxy)pyrimidin-4-yl]-methyl-amino]ethoxy]phenyl]methyl]-1,3-thiazolidine-2,4-dione, Nuclear receptor coactivator 1, Peroxisome proliferator-activated receptor gamma | | 著者 | Jang, J.Y, Han, B.W. | | 登録日 | 2017-09-07 | | 公開日 | 2018-09-12 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Structural Basis for the Enhanced Anti-Diabetic Efficacy of Lobeglitazone on PPAR gamma.
Sci Rep, 8, 2018
|
|
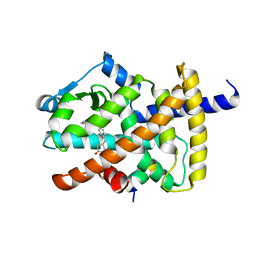
5YCP
 
 | |
7EC1
 
 | | Crystal structure of SdgB (ligand-free form) | | 分子名称: | GLYCEROL, Glycosyl transferase, group 1 family protein, ... | | 著者 | Kim, D.-G, Baek, I, Lee, Y, Kim, H.S. | | 登録日 | 2021-03-11 | | 公開日 | 2021-03-24 | | 最終更新日 | 2022-10-12 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural basis for SdgB- and SdgA-mediated glycosylation of staphylococcal adhesive proteins.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7EC6
 
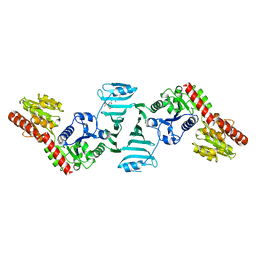 | | Crystal structure of SdgB (complexed with peptides) | | 分子名称: | ASP-SER-ASP, Glycosyl transferase, group 1 family protein | | 著者 | Kim, D.-G, Baek, I, Lee, Y, Kim, H.S. | | 登録日 | 2021-03-11 | | 公開日 | 2022-03-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural basis for SdgB- and SdgA-mediated glycosylation of staphylococcal adhesive proteins.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7EC7
 
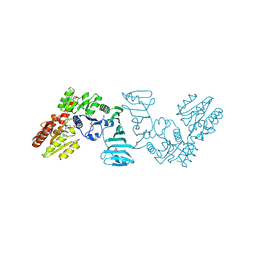 | | Crystal structure of SdgB (complexed with phosphate ions) | | 分子名称: | Glycosyl transferase, group 1 family protein, PHOSPHATE ION | | 著者 | Kim, D.-G, Baek, I, Lee, Y, Kim, H.S. | | 登録日 | 2021-03-11 | | 公開日 | 2022-03-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structural basis for SdgB- and SdgA-mediated glycosylation of staphylococcal adhesive proteins.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7EC3
 
 | | Crystal structure of SdgB (complexed with UDP, GlcNAc, and Glycosylated peptide) | | 分子名称: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-35)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-65)]5,6-DIHYDRO-BENZO[H]CINNOLIN-3-YLAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Glycosyl transferase, ... | | 著者 | Kim, D.-G, Baek, I, Lee, Y, Kim, H.S. | | 登録日 | 2021-03-11 | | 公開日 | 2022-03-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural basis for SdgB- and SdgA-mediated glycosylation of staphylococcal adhesive proteins.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
3SWE
 
 | | Haemophilus influenzae MurA in complex with UDP-N-acetylmuramic acid and covalent adduct of PEP with Cys117 | | 分子名称: | (2R)-2-{[(2R,3R,4R,5S,6R)-3-(acetylamino)-2-{[(S)-{[(R)-{[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}-5-hydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-4-yl]oxy}propanoic acid, GLYCEROL, SULFATE ION, ... | | 著者 | Zhu, J.-Y, Schonbrunn, E. | | 登録日 | 2011-07-13 | | 公開日 | 2012-03-14 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Functional Consequence of Covalent Reaction of Phosphoenolpyruvate with UDP-N-acetylglucosamine 1-Carboxyvinyltransferase (MurA).
J.Biol.Chem., 287, 2012
|
|
5LS0
 
 | | Crystal structure of Inorganic Pyrophosphatase PPA1 from Arabidopsis thaliana | | 分子名称: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, Soluble inorganic pyrophosphatase 1 | | 著者 | Grzechowiak, M, Sikorski, M, Jaskolski, M. | | 登録日 | 2016-08-22 | | 公開日 | 2017-09-13 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Crystal structures of plant inorganic pyrophosphatase, an enzyme with a moonlighting autoproteolytic activity.
Biochem.J., 476, 2019
|
|
6IJR
 
 | | Human PPARgamma ligand binding domain complexed with SB1495 | | 分子名称: | 16 mer peptide from Nuclear receptor coactivator 1, N-{[3-({[(1S,2R)-2-{[(2E)-2-cyano-4,4-dimethylpent-2-enoyl]amino}cyclopentyl]oxy}methyl)phenyl]methyl}-4-[(4-methylpiperazin-1-yl)methyl]benzamide, Peroxisome proliferator-activated receptor gamma | | 著者 | Jang, J.Y, Han, B.W. | | 登録日 | 2018-10-11 | | 公開日 | 2019-10-16 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Structural basis for the inhibitory effects of a novel reversible covalent ligand on PPAR gamma phosphorylation.
Sci Rep, 9, 2019
|
|
6IJS
 
 | | Human PPARgamma ligand binding domain complexed with SB1494 | | 分子名称: | 16-mer peptide from Nuclear receptor coactivator 1, N-{[3-({[(1R,2S)-2-{[(2E)-2-cyano-4,4-dimethylpent-2-enoyl]amino}cyclopentyl]oxy}methyl)phenyl]methyl}-4-[(4-methylpiperazin-1-yl)methyl]benzamide, Peroxisome proliferator-activated receptor gamma | | 著者 | Jang, J.Y, Han, B.W. | | 登録日 | 2018-10-11 | | 公開日 | 2019-10-16 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Structural basis for the inhibitory effects of a novel reversible covalent ligand on PPAR gamma phosphorylation.
Sci Rep, 9, 2019
|
|
6JQ7
 
 | |
7VFL
 
 | | Crystal structure of SdgB (UDP, NAG, and O-glycosylated SD peptide-binding form) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glycosyl transferase, group 1 family protein, ... | | 著者 | Kim, D.-G, Baek, I, Lee, Y, Kim, H.S. | | 登録日 | 2021-09-13 | | 公開日 | 2021-11-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Structural basis for SdgB- and SdgA-mediated glycosylation of staphylococcal adhesive proteins.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7VFO
 
 | | Crystal structure of SdgB (Phosphate-binding form) | | 分子名称: | Glycosyl transferase, group 1 family protein, PHOSPHATE ION | | 著者 | Kim, D.-G, Baek, I, Lee, Y, Kim, H.S. | | 登録日 | 2021-09-13 | | 公開日 | 2021-11-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structural basis for SdgB- and SdgA-mediated glycosylation of staphylococcal adhesive proteins.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7VFN
 
 | | Crystal structure of SdgB (SD peptide-binding form) | | 分子名称: | ASP-SER-ASP, Glycosyl transferase, group 1 family protein | | 著者 | Kim, D.-G, Baek, I, Lee, Y, Kim, H.S. | | 登録日 | 2021-09-13 | | 公開日 | 2021-11-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural basis for SdgB- and SdgA-mediated glycosylation of staphylococcal adhesive proteins.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7VFM
 
 | | Crystal structure of SdgB (UDP and SD peptide-binding form) | | 分子名称: | Glycosyl transferase, group 1 family protein, SER-ASP-SER-ASP, ... | | 著者 | Kim, D.-G, Baek, I, Lee, Y, Kim, H.S. | | 登録日 | 2021-09-13 | | 公開日 | 2021-11-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Structural basis for SdgB- and SdgA-mediated glycosylation of staphylococcal adhesive proteins.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
