8HF6
 
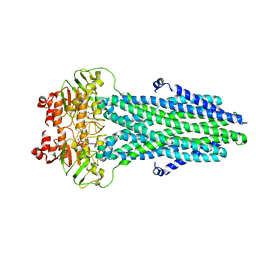 | | Cryo-EM structure of nucleotide-bound ComA E647Q mutant | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Competence factor transporting ATP-binding protein/permease ComA | | Authors: | Yu, L, Xin, X, Min, L. | | Deposit date: | 2022-11-09 | | Release date: | 2023-10-04 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of peptide secretion for Quorum sensing by ComA.
Nat Commun, 14, 2023
|
|
8HF7
 
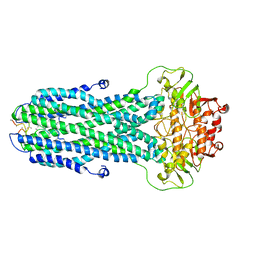 | |
4EYC
 
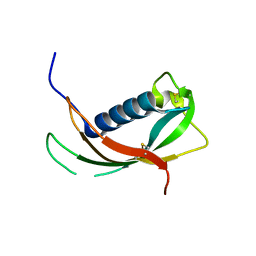 | | Crystal structure of the cathelin-like domain of human cathelicidin LL-37 (hCLD) | | Descriptor: | Cathelicidin antimicrobial peptide | | Authors: | Pazgier, M, Pozharski, E, Toth, E, Lu, W. | | Deposit date: | 2012-05-01 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Functional Analysis of the Pro-Domain of Human Cathelicidin, LL-37.
Biochemistry, 52, 2013
|
|
6KJ6
 
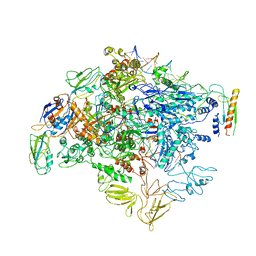 | | cryo-EM structure of Escherichia coli Crl transcription activation complex | | Descriptor: | DNA (51-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Xu, J, Zhang, Y. | | Deposit date: | 2019-07-21 | | Release date: | 2020-01-01 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Crl activates transcription by stabilizing active conformation of the master stress transcription initiation factor.
Elife, 8, 2019
|
|
7VKR
 
 | |
7VKK
 
 | | Crystal structure of D. melanogaster SAMTOR V66W/E67P mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, S-adenosylmethionine sensor upstream of mTORC1, SULFATE ION | | Authors: | Zhang, T, Ding, J. | | Deposit date: | 2021-09-30 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Molecular mechanism of S -adenosylmethionine sensing by SAMTOR in mTORC1 signaling.
Sci Adv, 8, 2022
|
|
7VKQ
 
 | |
5FJB
 
 | | Cyclophilin A Stabilize HIV-1 Capsid through a Novel Non- canonical Binding Site | | Descriptor: | GAG POLYPROTEIN, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE A | | Authors: | Liu, C, Perilla, J.R, Ning, J, Lu, M, Hou, G, Ramalhu, R, Bedwell, G.J, Ahn, J, Shi, J, Gronenborn, A.M, Prevelige Jr, P.E, Rousso, I, Aiken, C, Polenova, T, Schulten, K, Zhang, P. | | Deposit date: | 2015-10-07 | | Release date: | 2016-03-16 | | Last modified: | 2017-08-23 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Cyclophilin a Stabilizes the HIV-1 Capsid Through a Novel Non-Canonical Binding Site.
Nat.Commun., 7, 2016
|
|
6J0D
 
 | | Crystal structure of OsSUF4 | | Descriptor: | ZINC ION, transcription factor | | Authors: | Wang, B, Luo, Q. | | Deposit date: | 2018-12-24 | | Release date: | 2019-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The transcription factor OsSUF4 interacts with SDG725 in promoting H3K36me3 establishment.
Nat Commun, 10, 2019
|
|
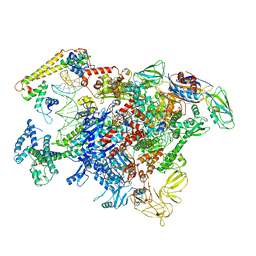
6LDI
 
 | |
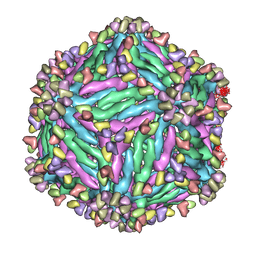
5Y0A
 
 | |
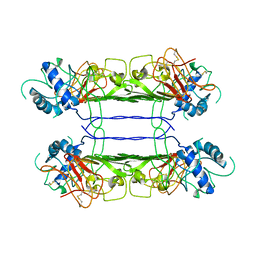
7WUT
 
 | |
7WUV
 
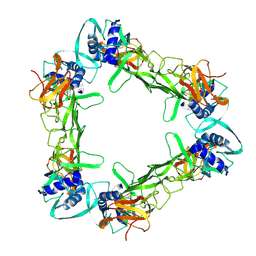 | | CryoEM structure of sNS1 hexamer | | Descriptor: | Core protein | | Authors: | Shu, B, Ooi, J.S.G, Lok, S.M. | | Deposit date: | 2022-02-09 | | Release date: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | CryoEM structures of the multimeric secreted NS1, a major factor for dengue hemorrhagic fever.
Nat Commun, 13, 2022
|
|
7WUR
 
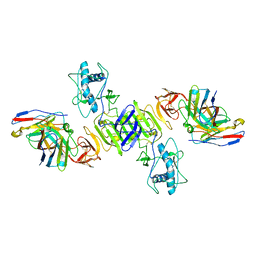 | | CryoEM structure of sNS1 complexed with Fab5E3 | | Descriptor: | Core protein, Fab 5E3 Heavy Chain, Fab 5E3 Light Chain | | Authors: | Shu, B, Lok, S.M. | | Deposit date: | 2022-02-09 | | Release date: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | CryoEM structures of the multimeric secreted NS1, a major factor for dengue hemorrhagic fever.
Nat Commun, 13, 2022
|
|
7WUS
 
 | |
7WUU
 
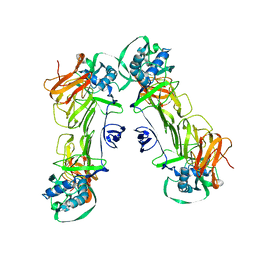 | | CryoEM structure of loose sNS1 tetramer | | Descriptor: | Core protein | | Authors: | Shu, B, Lok, S.M. | | Deposit date: | 2022-02-09 | | Release date: | 2022-12-21 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (8.3 Å) | | Cite: | CryoEM structures of the multimeric secreted NS1, a major factor for dengue hemorrhagic fever.
Nat Commun, 13, 2022
|
|
6A5F
 
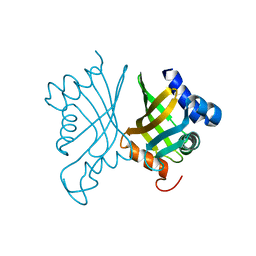 | |
6A5G
 
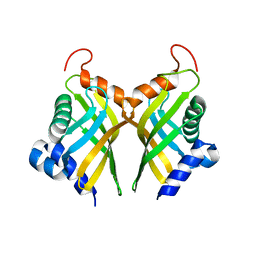 | |
5H32
 
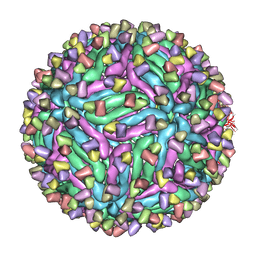 | | Cryo-EM structure of zika virus complexed with Fab C10 at pH 5.0 | | Descriptor: | C10 IgG heavy chain variable region, C10 IgG light chain variable region, structural protein E | | Authors: | Zhang, S, Kostyuchenko, V, Ng, T.-S, Lok, S.-M. | | Deposit date: | 2016-10-20 | | Release date: | 2016-11-30 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Neutralization mechanism of a highly potent antibody against Zika virus
Nat Commun, 7, 2016
|
|
5H30
 
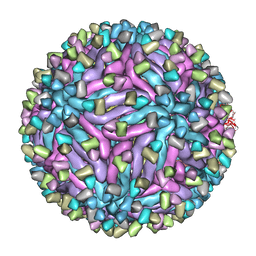 | | Cryo-EM structure of zika virus complexed with Fab C10 at pH 6.5 | | Descriptor: | IgG C10 heavy chain, IgG C10 light chain, structural protein E, ... | | Authors: | Zhang, S, Kostyuchenko, V, Ng, T.-S, Lok, S.-M. | | Deposit date: | 2016-10-19 | | Release date: | 2016-11-30 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Neutralization mechanism of a highly potent antibody against Zika virus
Nat Commun, 7, 2016
|
|
6AY9
 
 | | Structure of the native full-length HIV-1 capsid protein in complex with CPSF6 peptide | | Descriptor: | CHLORIDE ION, Cleavage and polyadenylation specificity factor subunit 6, HIV-1 capsid protein, ... | | Authors: | Gres, A.T, Kirby, K.A, Sarafianos, S.G. | | Deposit date: | 2017-09-07 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Multidisciplinary studies with mutated HIV-1 capsid proteins reveal structural mechanisms of lattice stabilization.
Nat Commun, 14, 2023
|
|
6B2K
 
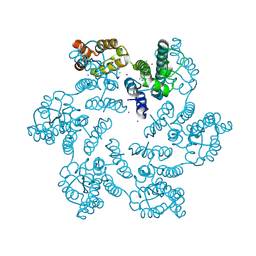 | | E45A/R132T mutant of HIV-1 capsid protein | | Descriptor: | CHLORIDE ION, HIV-1 capsid protein, IODIDE ION | | Authors: | Gres, A.T, Kirby, K.A, Sarafianos, S.G. | | Deposit date: | 2017-09-20 | | Release date: | 2018-09-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Multidisciplinary studies with mutated HIV-1 capsid proteins reveal structural mechanisms of lattice stabilization.
Nat Commun, 14, 2023
|
|
6AYA
 
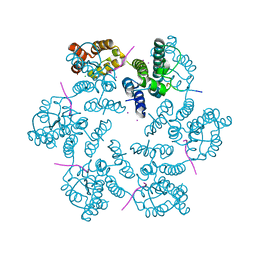 | | Structure of the native full-length HIV-1 capsid protein in complex with Nup153 peptide | | Descriptor: | CHLORIDE ION, HIV-1 capsid protein, IODIDE ION, ... | | Authors: | Gres, A.T, Kirby, K.A, Sarafianos, S.G. | | Deposit date: | 2017-09-07 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Multidisciplinary studies with mutated HIV-1 capsid proteins reveal structural mechanisms of lattice stabilization.
Nat Commun, 14, 2023
|
|
6B2J
 
 | |
6B2I
 
 | | E45A mutant of the HIV-1 capsid protein | | Descriptor: | CHLORIDE ION, HIV-1 capsid protein, IODIDE ION | | Authors: | Gres, A.T, Kirby, K.A, Sarafianos, S.G. | | Deposit date: | 2017-09-20 | | Release date: | 2018-09-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Multidisciplinary studies with mutated HIV-1 capsid proteins reveal structural mechanisms of lattice stabilization.
Nat Commun, 14, 2023
|
|
