5JSG
 
 | | Crystal structure of Spindlin1 bound to compound EML405 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Spindlin-1, ... | | Authors: | Su, X, Li, H. | | Deposit date: | 2016-05-08 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Developing Spindlin1 small-molecule inhibitors by using protein microarrays
Nat. Chem. Biol., 13, 2017
|
|
5JSJ
 
 | | Crystal structure of Spindlin1 bound to compound EML631 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Spindlin-1, ... | | Authors: | Su, X, Li, H. | | Deposit date: | 2016-05-08 | | Release date: | 2017-05-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.348 Å) | | Cite: | Developing Spindlin1 small-molecule inhibitors by using protein microarrays
Nat. Chem. Biol., 13, 2017
|
|
6YMV
 
 | |
6YMW
 
 | |
2PE2
 
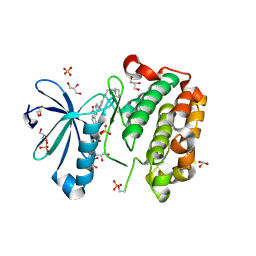 | |
3HHU
 
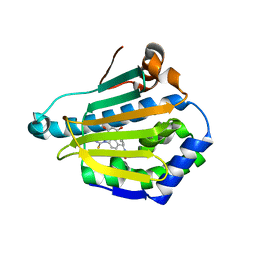 | | Human heat-shock protein 90 (HSP90) in complex with {4-[3-(2,4-dihydroxy-5-isopropyl-phenyl)-5-thioxo- 1,5-dihydro-[1,2,4]triazol-4-yl]-benzyl}-carbamic acid ethyl ester {ZK 2819} | | Descriptor: | Heat shock protein HSP 90-alpha, ethyl (4-{3-[2,4-dihydroxy-5-(1-methylethyl)phenyl]-5-sulfanyl-4H-1,2,4-triazol-4-yl}benzyl)carbamate | | Authors: | Adler, M, Whitlow, M. | | Deposit date: | 2009-05-17 | | Release date: | 2009-07-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Potent triazolothione inhibitor of heat-shock protein-90.
Chem.Biol.Drug Des., 74, 2009
|
|
6IZQ
 
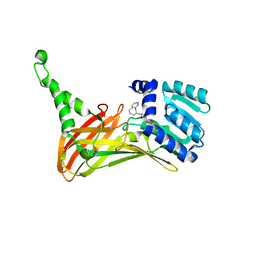 | | PRMT4 bound with a bicyclic compound | | Descriptor: | (2R)-1-(methylamino)-3-(1,3,4,5-tetrahydro-2-benzazepin-2-yl)propan-2-ol, Histone-arginine methyltransferase CARM1 | | Authors: | Xiong, B, Cao, D.Y, Guo, Z.H, Li, Y.L, Li, J, Huang, X, Shen, J.K. | | Deposit date: | 2018-12-20 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.449 Å) | | Cite: | Design and Synthesis of Potent, Selective Inhibitors of Protein Arginine Methyltransferase 4 against Acute Myeloid Leukemia.
J.Med.Chem., 62, 2019
|
|
4EZ5
 
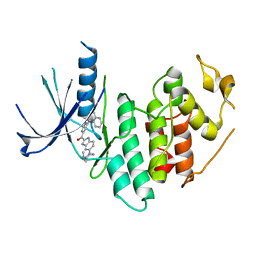 | | CDK6 (monomeric) in complex with inhibitor | | Descriptor: | Cyclin-dependent kinase 6, {5-[4-(dimethylamino)piperidin-1-yl]-1H-imidazo[4,5-b]pyridin-2-yl}[2-(isoquinolin-4-yl)pyridin-4-yl]methanone | | Authors: | Chopra, R, Xu, M. | | Deposit date: | 2012-05-02 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Fragment-Based Discovery of 7-Azabenzimidazoles as Potent, Highly Selective, and Orally Active CDK4/6 Inhibitors.
ACS Med Chem Lett, 3, 2012
|
|
4G2Y
 
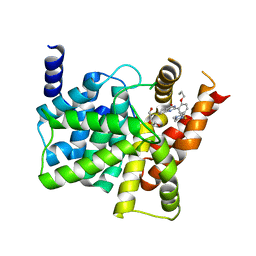 | | Crystal structure of PDE5A complexed with its inhibitor | | Descriptor: | 2-{5-[(4-methylpiperazin-1-yl)sulfonyl]-2-propoxyphenyl}-3,5,6,7-tetrahydro-4H-cyclopenta[d]pyrimidin-4-one, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Ren, J, Chen, T.T, Xu, Y.C. | | Deposit date: | 2012-07-13 | | Release date: | 2013-06-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design, synthesis, and pharmacological evaluation of monocyclic pyrimidinones as novel inhibitors of PDE5.
J.Med.Chem., 55, 2012
|
|
8IJK
 
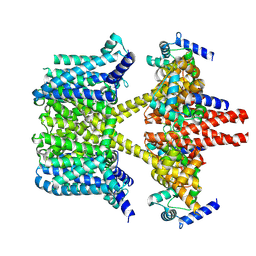 | | human KCNQ2-CaM-Ebio1 complex in the presence of PIP2 | | Descriptor: | Calmodulin-1, N-(1,2-dihydroacenaphthylen-5-yl)-4-fluoranyl-benzamide, Potassium voltage-gated channel subfamily KQT member 2 | | Authors: | Ma, D, Guo, J. | | Deposit date: | 2023-02-27 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A small-molecule activation mechanism that directly opens the KCNQ2 channel.
Nat.Chem.Biol., 2024
|
|
7DFH
 
 | |
7DFG
 
 | |
7DOI
 
 | |
7DOK
 
 | |
4G2W
 
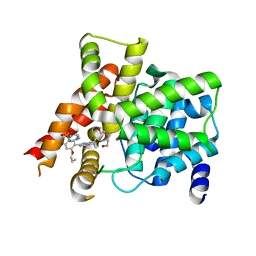 | | Crystal structure of PDE5A in complex with its inhibitor | | Descriptor: | 5,6-diethyl-2-{5-[(4-methylpiperazin-1-yl)sulfonyl]-2-propoxyphenyl}pyrimidin-4(3H)-one, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Ren, J, Chen, T.T, Xu, Y.C. | | Deposit date: | 2012-07-13 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Design, synthesis, and pharmacological evaluation of monocyclic pyrimidinones as novel inhibitors of PDE5.
J.Med.Chem., 55, 2012
|
|
4IA0
 
 | | Crystal structure of the PDE5A1 catalytic domain in complex with novel inhibitors | | Descriptor: | 5-bromo-2-{2-ethoxy-5-[(4-methylpiperazin-1-yl)sulfonyl]phenyl}-6-octylpyrimidin-4(3H)-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Ren, J, Chen, T, Xu, Y. | | Deposit date: | 2012-12-05 | | Release date: | 2014-01-01 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Exploration of the 5-bromopyrimidin-4(3H)-ones as potent inhibitors of PDE5.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4I9Z
 
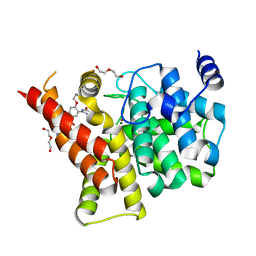 | | Crystal structure of the PDE5A1 catalytic domain in complex with novel inhibitors | | Descriptor: | 5-bromo-2-{5-[(4-methylpiperazin-1-yl)acetyl]-2-propoxyphenyl}-6-(propan-2-yl)pyrimidin-4(3H)-one, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Ren, J, Chen, T, Xu, Y. | | Deposit date: | 2012-12-05 | | Release date: | 2014-01-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Exploration of the 5-bromopyrimidin-4(3H)-ones as potent inhibitors of PDE5.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3SIE
 
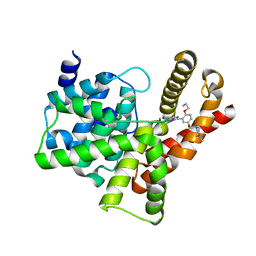 | | Crystal structure of the PDE5A1 catalytic domain in complex with novel inhibitors | | Descriptor: | 5-bromo-6-ethyl-2-{5-[(4-methylpiperazin-1-yl)sulfonyl]-2-propoxyphenyl}pyrimidin-4(3H)-one, cGMP-specific 3',5'-cyclic phosphodiesterase | | Authors: | Chen, T.T, Chen, T, Xu, Y.C. | | Deposit date: | 2011-06-17 | | Release date: | 2011-08-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Utilization of Halogen Bond in Lead Optimization: A Case Study of Rational Design of Potent Phosphodiesterase Type 5 (PDE5) Inhibitors.
J.Med.Chem., 54, 2011
|
|
8JKD
 
 | |
8JLW
 
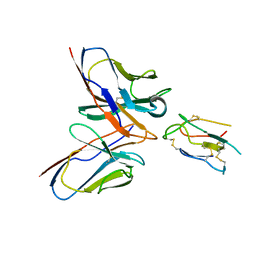 | | CCHFV envelope protein Gc in complex with Gc8 | | Descriptor: | Glycoprotein C,CCHFV envelope protein Gc fusion loops, Mouse antibody Gc8 heavy chain, Mouse antibody Gc8 light chain | | Authors: | Chong, T, Cao, S. | | Deposit date: | 2023-06-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Neutralizing monoclonal antibodies against the Gc fusion loop region of Crimean-Congo hemorrhagic fever virus.
Plos Pathog., 20, 2024
|
|
8JLX
 
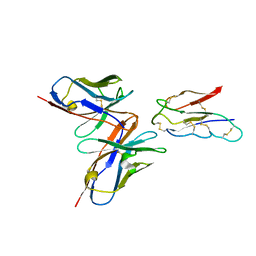 | | CCHFV envelope protein Gc in complex with Gc13 | | Descriptor: | Glycoprotein C,CCHFV Gc fusion loops, Mouse antibody Gc13 heavy chain, Mouse antibody Gc13 light chain | | Authors: | Chong, T, Cao, S. | | Deposit date: | 2023-06-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Neutralizing monoclonal antibodies against the Gc fusion loop region of Crimean-Congo hemorrhagic fever virus.
Plos Pathog., 20, 2024
|
|
7TAI
 
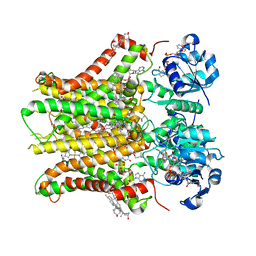 | | Structure of STEAP2 in complex with ligands | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, CHOLESTEROL, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Wang, L, Chen, K.H, Zhou, M. | | Deposit date: | 2021-12-20 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of stepwise electron transfer in six-transmembrane epithelial antigen of the prostate (STEAP) 1 and 2.
Elife, 12, 2023
|
|
7TTI
 
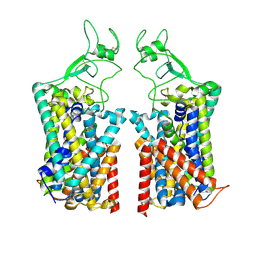 | | Human KCC1 bound with VU0463271 In an outward-open state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-cyclopropyl-N-(4-methyl-1,3-thiazol-2-yl)-2-[(6-phenylpyridazin-3-yl)sulfanyl]acetamide, Solute carrier family 12 member 4 | | Authors: | Zhao, Y.X, Cao, E.H. | | Deposit date: | 2022-02-01 | | Release date: | 2022-06-29 | | Last modified: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the human cation-chloride cotransport KCC1 in an outward-open state.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7TTH
 
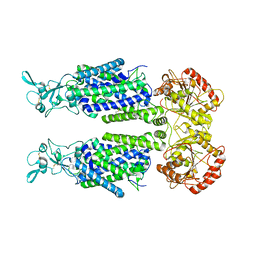 | | Human potassium-chloride cotransporter 1 in inward-open state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, POTASSIUM ION, Solute carrier family 12 member 4 | | Authors: | Zhao, Y.X, Cao, E.H. | | Deposit date: | 2022-02-01 | | Release date: | 2022-06-29 | | Last modified: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structure of the human cation-chloride cotransport KCC1 in an outward-open state.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7TWD
 
 | | Structure of AAGAB C-terminal dimerization domain | | Descriptor: | Alpha- and gamma-adaptin-binding protein p34, PHOSPHATE ION | | Authors: | Tian, Y, Yin, Q. | | Deposit date: | 2022-02-07 | | Release date: | 2023-01-18 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Oligomer-to-monomer transition underlies the chaperone function of AAGAB in AP1/AP2 assembly.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
