8C1U
 
 | | SFX structure of D.m(6-4)photolyase | | Descriptor: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Cellini, A, Kumar, M, Nimmrich, A, Mutisya, J, Furrer, A, Beale, E.V, Carrillo, M, Malla, T.N, Maj, P, Dworkowskic, F, Cirelli, C, Ozerovi, D, Bacellar, C, Strandfuss, J, Weinert, T, Ihalainen, J.A, Yuan Wahlgren, W, Westenhoff, S. | | Deposit date: | 2022-12-21 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Femtosecond serial crystallography resolves conformational changes which guide long-range electron transfer in a photolyase
To Be Published
|
|
8C69
 
 | | Light SFX structure of D.m(6-4)photolyase at 100 microsecond time delay | | Descriptor: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Cellini, A, Kumar, M, Nimmrich, A, Mutisya, J, Furrer, A, Beale, E.V, Carrillo, M, Malla, T.N, Maj, P, Dworkowskic, F, Cirelli, C, Ozerovi, D, Bacellar, C, Strandfuss, J, Weinert, T, Ihalainen, J.A, Yuan Wahlgren, W, Westenhoff, S. | | Deposit date: | 2023-01-11 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Femtosecond serial crystallography resolves conformational changes which guide long-range electron transfer in a photolyase
To Be Published
|
|
8C6A
 
 | | Light SFX structure of D.m(6-4)photolyase at 1ps time delay | | Descriptor: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Cellini, A, Kumar, M, Nimmrich, A, Mutisya, J, Furrer, A, Beale, E.V, Carrillo, M, Malla, T.N, Maj, P, Dworkowskic, F, Cirelli, C, Ozerovi, D, Bacellar, C, Strandfuss, J, Weinert, T, Ihalainen, J.A, Yuan Wahlgren, W, Westenhoff, S. | | Deposit date: | 2023-01-11 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Femtosecond serial crystallography resolves conformational changes which guide long-range electron transfer in a photolyase
To Be Published
|
|
8C6B
 
 | | Light SFX structure of D.m(6-4)photolyase at 20ps time delay | | Descriptor: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Cellini, A, Kumar, M, Nimmrich, A, Mutisya, J, Furrer, A, Beale, E.V, Carrillo, M, Malla, T.N, Maj, P, Dworkowskic, F, Cirelli, C, Ozerovi, D, Bacellar, C, Strandfuss, J, Weinert, T, Ihalainen, J.A, Yuan Wahlgren, W, Westenhoff, S. | | Deposit date: | 2023-01-11 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Femtosecond serial crystallography resolves conformational changes which guide long-range electron transfer in a photolyase
To Be Published
|
|
8C6F
 
 | | Light SFX structure of D.m(6-4)photolyase at 400fs time delay | | Descriptor: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Cellini, A, Kumar, M, Nimmrich, A, Mutisya, J, Furrer, A, Beale, E.V, Carrillo, M, Malla, T.N, Maj, P, Dworkowskic, F, Cirelli, C, Ozerovi, D, Bacellar, C, Strandfuss, J, Weinert, T, Ihalainen, J.A, Yuan Wahlgren, W, Westenhoff, S. | | Deposit date: | 2023-01-11 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Femtosecond serial crystallography resolves conformational changes which guide long-range electron transfer in a photolyase
To Be Published
|
|
6TI7
 
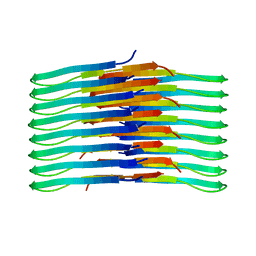 | | Mixing Abeta(1-40) and Abeta(1-42) peptides generates unique amyloid fibrils | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Cerofolini, L, Ravera, E, Bologna, S, Wiglenda, T, Boddrich, A, Purfurst, B, Benilova, A, Korsak, M, Gallo, G, Rizzo, D, Gonnelli, L, Fragai, M, De Strooper, B, Wanker, E.E, Luchinat, C. | | Deposit date: | 2019-11-21 | | Release date: | 2020-07-22 | | Last modified: | 2023-06-14 | | Method: | SOLID-STATE NMR | | Cite: | Mixing A beta (1-40) and A beta (1-42) peptides generates unique amyloid fibrils.
Chem.Commun.(Camb.), 56, 2020
|
|
6TI6
 
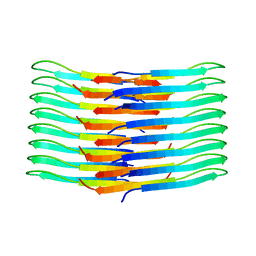 | | Mixing Abeta(1-40) and Abeta(1-42) peptides generates unique amyloid fibrils | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Cerofolini, L, Ravera, E, Bologna, S, Wiglenda, T, Boddrich, A, Purfurst, B, Benilova, A, Korsak, M, Gallo, G, Rizzo, D, Gonnelli, L, Fragai, M, De Strooper, B, Wanker, E.E, Luchinat, C. | | Deposit date: | 2019-11-21 | | Release date: | 2020-07-22 | | Last modified: | 2023-06-14 | | Method: | SOLID-STATE NMR | | Cite: | Mixing A beta (1-40) and A beta (1-42) peptides generates unique amyloid fibrils.
Chem.Commun.(Camb.), 56, 2020
|
|
6YO5
 
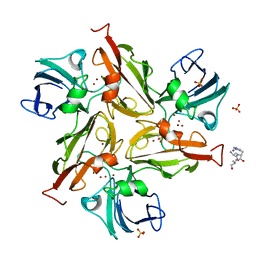 | | Crystal structure of the M295F variant of Ssl1 | | Descriptor: | ALA-HIS-ALA, COPPER (II) ION, Copper oxidase, ... | | Authors: | Mielenbrink, S, Olbrich, A, Urlacher, V, Span, I. | | Deposit date: | 2020-04-14 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Effect of the axial ligand mutation on spectral and structural properties of Ssl1 laccase
To Be Published
|
|
6Y4A
 
 | |
6YZF
 
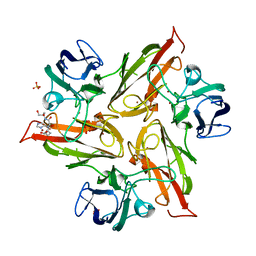 | | Crystal structure of the M295Y variant of Ssl1 | | Descriptor: | COPPER (II) ION, Copper oxidase, GLU-HIS-SER, ... | | Authors: | Mielenbrink, S, Olbrich, A, Urlacher, V, Span, I. | | Deposit date: | 2020-05-06 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.684 Å) | | Cite: | Effect of the axial ligand mutation on spectral and structural properties of Ssl1 laccase
To Be Published
|
|
6YZD
 
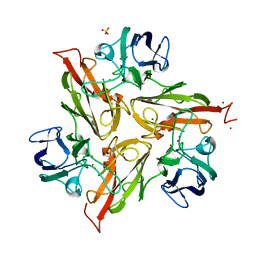 | |
6YZY
 
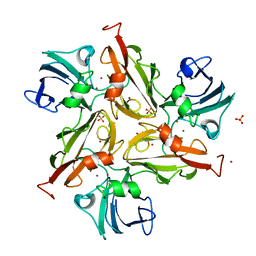 | |
2C5D
 
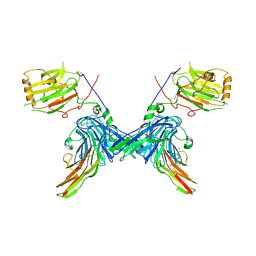 | | Structure of a minimal Gas6-Axl complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GROWTH-ARREST-SPECIFIC PROTEIN 6 PRECURSOR, ... | | Authors: | Sasaki, T, Knyazev, P.G, Clout, N.J, Cheburkin, Y, Goehring, W, Ullrich, A, Timpl, R, Hohenester, E. | | Deposit date: | 2005-10-26 | | Release date: | 2005-12-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural Basis for Gas6-Axl Signalling.
Embo J., 25, 2006
|
|
1N4G
 
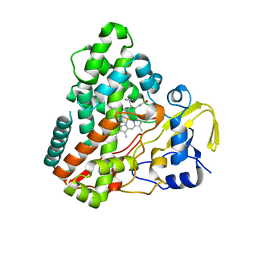 | | Structure of CYP121, a Mycobacterial P450, in Complex with Iodopyrazole | | Descriptor: | 4-IODOPYRAZOLE, Cytochrome P450 121, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Leys, D, Mowat, C.G, McLean, K.J, Richmond, A, Chapman, S.K, Walkinshaw, M.D, Munro, A.W, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2002-10-31 | | Release date: | 2003-02-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Atomic structure of Mycobacterium tuberculosis CYP121 to 1.06 A reveals novel features of cytochrome P450.
J.Biol.Chem., 278, 2003
|
|
1H30
 
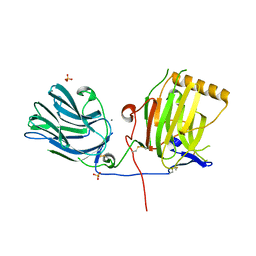 | | C-terminal LG domain pair of human Gas6 | | Descriptor: | CALCIUM ION, GROWTH-ARREST-SPECIFIC PROTEIN, SULFATE ION | | Authors: | Sasaki, T, Knyazev, P.G, Cheburkin, Y, Gohring, W, Tisi, D, Ullrich, A, Timpl, R, Hohenester, E. | | Deposit date: | 2002-08-21 | | Release date: | 2003-01-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of a Carboxy-Terminal Fragment of Growth-Arrest-Specific Protein Gas6: Receptor Tyrosine Kinase Activation by Laminin G-Like Domains
J.Biol.Chem., 277, 2002
|
|
1N40
 
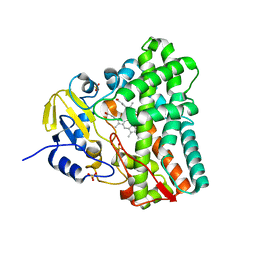 | | Atomic structure of CYP121, a mycobacterial P450 | | Descriptor: | Cytochrome P450 121, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Leys, D, Mowat, C.G, McLean, K.J, Richmond, A, Chapman, S.K, Walkinshaw, M.D, Munro, A.W, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2002-10-30 | | Release date: | 2003-02-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Atomic structure of Mycobacterium tuberculosis CYP121 to 1.06 A reveals novel features of cytochrome P450.
J.Biol.Chem., 278, 2003
|
|
2VAQ
 
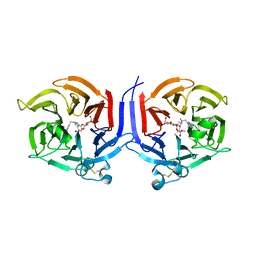 | | STRUCTURE OF STRICTOSIDINE SYNTHASE IN COMPLEX WITH INHIBITOR | | Descriptor: | (2S,3R,4S)-methyl 4-(2-(2-(1H-indol-3-yl)ethylamino)ethyl)-2-((2S,3R,4S,5S,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yloxy)-3-vinyl-3,4-dihydro-2H-pyran-5-carboxylate, STRICTOSIDINE SYNTHASE | | Authors: | Maresh, J, Giddings, L.A, Friedrich, A, Loris, E.A, Panjikar, S, Trout, B.L, Stoeckigt, J, Peters, B, O'Connor, S.E. | | Deposit date: | 2007-09-04 | | Release date: | 2008-09-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Strictosidine synthase: mechanism of a Pictet-Spengler catalyzing enzyme.
J. Am. Chem. Soc., 130, 2008
|
|
4PED
 
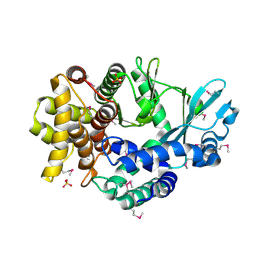 | | Mitochondrial ADCK3 employs an atypical protein kinase-like fold to enable coenzyme Q biosynthes | | Descriptor: | Chaperone activity of bc1 complex-like, mitochondrial, SULFATE ION | | Authors: | Bingman, C.A, Smith, R, Joshi, S, Stefely, J.A, Reidenbach, A.G, Ulbrich, A, Oruganty, O, Floyd, B.J, Jochem, A, Saunders, J.M, Johnson, I.E, Wrobel, R.L, Barber, G.E, Lee, D, Li, S, Kannan, N, Coon, J.J, Pagliarini, D.J, Mitochondrial Protein Partnership (MPP) | | Deposit date: | 2014-04-22 | | Release date: | 2014-11-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Mitochondrial ADCK3 Employs an Atypical Protein Kinase-like Fold to Enable Coenzyme Q Biosynthesis.
Mol.Cell, 57, 2015
|
|
1UUM
 
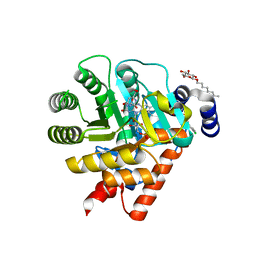 | | Rat dihydroorotate dehydrogenase (DHOD)in complex with atovaquone | | Descriptor: | 2-[4-(4-CHLOROPHENYL)CYCLOHEXYLIDENE]-3,4-DIHYDROXY-1(2H)-NAPHTHALENONE, DIHYDROOROTATE DEHYDROGENASE, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Hansen, M, Le Nours, J, Johansson, E, Antal, T, Ullrich, A, Loffler, M, Larsen, S. | | Deposit date: | 2004-01-06 | | Release date: | 2004-04-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Inhibitor Binding in a Class 2 Dihydroorotate Dehydrogenase Causes Variations in the Membrane-Associated N-Terminal Domain
Protein Sci., 13, 2004
|
|
1UUO
 
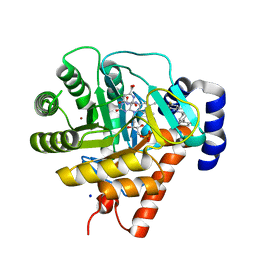 | | Rat dihydroorotate dehydrogenase (DHOD)in complex with brequinar | | Descriptor: | 6-FLUORO-2-(2'-FLUORO-1,1'-BIPHENYL-4-YL)-3-METHYLQUINOLINE-4-CARBOXYLIC ACID, DIHYDROOROTATE DEHYDROGENASE, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Hansen, M, Le Nours, J, Johansson, E, Antal, T, Ullrich, A, Loffler, M, Larsen, S. | | Deposit date: | 2004-01-08 | | Release date: | 2004-04-01 | | Last modified: | 2019-01-30 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Inhibitor Binding in a Class 2 Dihydroorotate Dehydrogenase Causes Variations in the Membrane-Associated N-Terminal Domain
Protein Sci., 13, 2004
|
|
2GVX
 
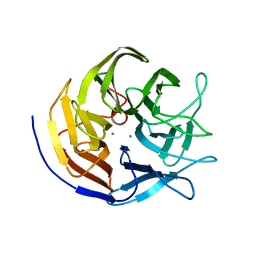 | | Structure of diisopropyl fluorophosphatase (DFPase), mutant D229N / N175D | | Descriptor: | CALCIUM ION, diisopropyl fluorophosphatase | | Authors: | Blum, M.-M, Lohr, F, Richardt, A, Ruterjans, H, Chen, J.C.-H. | | Deposit date: | 2006-05-03 | | Release date: | 2006-09-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding of a Designed Substrate Analogue to Diisopropyl Fluorophosphatase: Implications for the Phosphotriesterase Mechanism
J.Am.Chem.Soc., 128, 2006
|
|
6HF3
 
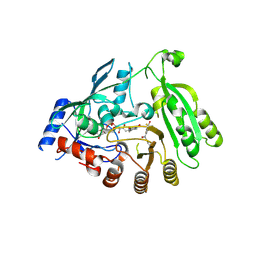 | | M tuberculosis DprE1 in complex with a covalently bound nitrobenzothiazinone | | Descriptor: | 2-(4-(cyclohexylmethyl)piperazin-1-yl)-8-nitro-6-(trifluoromethyl)-4H-benzo[e][1,3]thiazin-4-one, bound form, Decaprenylphosphoryl-beta-D-ribose oxidase, ... | | Authors: | Futterer, K, Batt, S.M, Besra, G.S. | | Deposit date: | 2018-08-21 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Novel insight into the reaction of nitro, nitroso and hydroxylamino benzothiazinones and of benzoxacinones with Mycobacterium tuberculosis DprE1.
Sci Rep, 8, 2018
|
|
6HEZ
 
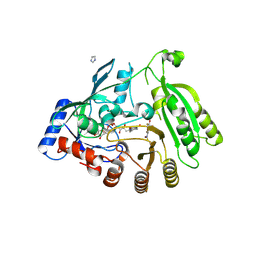 | | M tuberculosis DprE1 in complex with BTZ043 | | Descriptor: | 8-(hydroxyamino)-2-[(2S)-2-methyl-1,4-dioxa-8-azaspiro[4.5]dec-8-yl]-6-(trifluoromethyl)-4H-1,3-benzothiazin-4-one, Decaprenylphosphoryl-beta-D-ribose oxidase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Futterer, K, Batt, S.M, Besra, G.S. | | Deposit date: | 2018-08-21 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Novel insight into the reaction of nitro, nitroso and hydroxylamino benzothiazinones and of benzoxacinones with Mycobacterium tuberculosis DprE1.
Sci Rep, 8, 2018
|
|
6HFW
 
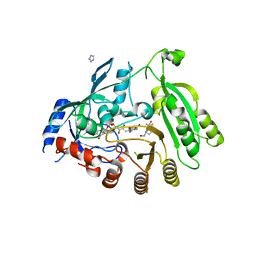 | | Mycobacterium tuberculosis DprE1 in complex with CMP1 | | Descriptor: | 8-(oxidanylamino)-2-piperidin-1-yl-6-(trifluoromethyl)-1,3-benzothiazin-4-one, Decaprenylphosphoryl-beta-D-ribose oxidase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Chung, C. | | Deposit date: | 2018-08-21 | | Release date: | 2018-09-19 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Novel insight into the reaction of nitro, nitroso and hydroxylamino benzothiazinones and of benzoxacinones with Mycobacterium tuberculosis DprE1.
Sci Rep, 8, 2018
|
|
6HFV
 
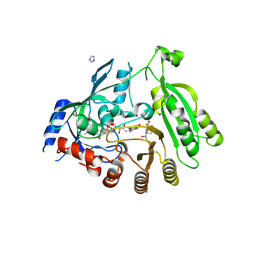 | | Mycobacterium tuberculosis DprE1 in complex with CMP2 | | Descriptor: | 8-(oxidanylamino)-2-piperidin-1-yl-6-(trifluoromethyl)-1,3-benzothiazin-4-one, Decaprenylphosphoryl-beta-D-ribose oxidase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Chung, C. | | Deposit date: | 2018-08-21 | | Release date: | 2018-09-19 | | Last modified: | 2019-08-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Novel insight into the reaction of nitro, nitroso and hydroxylamino benzothiazinones and of benzoxacinones with Mycobacterium tuberculosis DprE1.
Sci Rep, 8, 2018
|
|
