6YZP
 
 | | Carborane nido-hexyl-sulfonamide in complex with CA IX mimic | | Descriptor: | Carbonic anhydrase 2, Carborane nido-hexyl-sulfonamide, ZINC ION | | Authors: | Kugler, M, Brynda, J, Pospisilova, K, Rezacova, P. | | Deposit date: | 2020-05-07 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The structural basis for the selectivity of sulfonamido dicarbaboranes toward cancer-associated carbonic anhydrase IX.
J Enzyme Inhib Med Chem, 35, 2020
|
|
6YZV
 
 | | Closo-carborane ethyl-sulfonamide in complex with CA II | | Descriptor: | Carbonic anhydrase 2, Carborane ethyl-sulfonamide, ZINC ION | | Authors: | Kugler, M, Brynda, J, Pospisilova, K, Rezacova, P. | | Deposit date: | 2020-05-07 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The structural basis for the selectivity of sulfonamido dicarbaboranes toward cancer-associated carbonic anhydrase IX.
J Enzyme Inhib Med Chem, 35, 2020
|
|
5O46
 
 | | Crystal structure of Iristatin, a secreted salivary cystatin from the hard tick Ixodes ricinus | | Descriptor: | GLYCEROL, Iristatin | | Authors: | Busa, M, Rezacova, P, Kotal, J, Kotsyfakis, M, Mares, M. | | Deposit date: | 2017-05-26 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | The structure and function of Iristatin, a novel immunosuppressive tick salivary cystatin.
Cell.Mol.Life Sci., 76, 2019
|
|
3I8W
 
 | |
5YI9
 
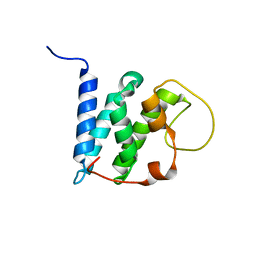 | | Solution structure of the LEDGF/p75 IBD - JPO2 (aa 56-91) complex | | Descriptor: | PC4 and SFRS1-interacting protein,Cell division cycle-associated 7-like protein | | Authors: | Veverka, V. | | Deposit date: | 2017-10-03 | | Release date: | 2018-08-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Affinity switching of the LEDGF/p75 IBD interactome is governed by kinase-dependent phosphorylation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1CL7
 
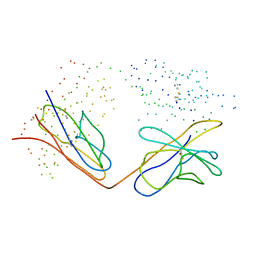 | | ANTI HIV1 PROTEASE FAB | | Descriptor: | PROTEIN (IGG1 ANTIBODY 1696 (constant heavy chain)), PROTEIN (IGG1 ANTIBODY 1696 (light chain)), PROTEIN (IGG1 ANTIBODY 1696 (variable heavy chain)) | | Authors: | Lescar, J, Bentley, G.A. | | Deposit date: | 1999-05-06 | | Release date: | 2000-01-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Inhibition of the HIV-1 and HIV-2 proteases by a monoclonal antibody.
Protein Sci., 8, 1999
|
|
6SYI
 
 | |
6R5H
 
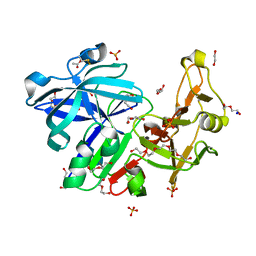 | | Major aspartyl peptidase 1 from C. neoformans | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, ... | | Authors: | Krystufek, R, Sacha, P, Brynda, J, Konvalinka, J. | | Deposit date: | 2019-03-25 | | Release date: | 2021-04-07 | | Last modified: | 2021-06-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Re-emerging Aspartic Protease Targets: Examining Cryptococcus neoformans Major Aspartyl Peptidase 1 as a Target for Antifungal Drug Discovery.
J.Med.Chem., 64, 2021
|
|
6R6A
 
 | | Major aspartyl peptidase 1 from C. neoformans | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, Endopeptidase, ... | | Authors: | Krystufek, R, Sacha, P, Brynda, J, Konvalinka, J. | | Deposit date: | 2019-03-26 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Re-emerging Aspartic Protease Targets: Examining Cryptococcus neoformans Major Aspartyl Peptidase 1 as a Target for Antifungal Drug Discovery.
J.Med.Chem., 64, 2021
|
|
6EMO
 
 | | Solution structure of the LEDGF/p75 IBD - JPO2 (aa 1-32) complex | | Descriptor: | PC4 and SFRS1-interacting protein,LEDGF/p75 IBD-JPO2 M1 | | Authors: | Veverka, V. | | Deposit date: | 2017-10-03 | | Release date: | 2018-07-25 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Affinity switching of the LEDGF/p75 IBD interactome is governed by kinase-dependent phosphorylation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6EMP
 
 | | Solution structure of the LEDGF/p75 IBD - POGZ (aa 1370-1404) complex | | Descriptor: | PC4 and SFRS1-interacting protein,Pogo transposable element with ZNF domain | | Authors: | Veverka, V. | | Deposit date: | 2017-10-03 | | Release date: | 2018-07-25 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Affinity switching of the LEDGF/p75 IBD interactome is governed by kinase-dependent phosphorylation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6EMQ
 
 | | Solution structure of the LEDGF/p75 IBD - MLL1 (aa 111-160) complex | | Descriptor: | PC4 and SFRS1-interacting protein,Histone-lysine N-methyltransferase 2A | | Authors: | Veverka, V. | | Deposit date: | 2017-10-03 | | Release date: | 2018-08-01 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Affinity switching of the LEDGF/p75 IBD interactome is governed by kinase-dependent phosphorylation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6EMR
 
 | |
6G01
 
 | |
4O1G
 
 | | MTB adenosine kinase in complex with gamma-Thio-ATP | | Descriptor: | Adenosine kinase, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, SODIUM ION | | Authors: | Dostal, J, Brynda, J, Hocek, M, Pichova, I. | | Deposit date: | 2013-12-15 | | Release date: | 2014-11-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis for Inhibition of Mycobacterial and Human Adenosine Kinase by 7-Substituted 7-(Het)aryl-7-deazaadenine Ribonucleosides
J.Med.Chem., 57, 2014
|
|
4O1L
 
 | | Human Adenosine Kinase in complex with inhibitor | | Descriptor: | 5-ethynyl-7-(beta-D-ribofuranosyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine, Adenosine kinase, MAGNESIUM ION | | Authors: | Brynda, J, Dostal, J, Pichova, I, Hodcek, M. | | Deposit date: | 2013-12-16 | | Release date: | 2014-11-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Inhibition of Mycobacterial and Human Adenosine Kinase by 7-Substituted 7-(Het)aryl-7-deazaadenine Ribonucleosides
J.Med.Chem., 57, 2014
|
|
3PO6
 
 | | Crystal structure of human carbonic anhydrase II with 6,7-Dimethoxy-1-methyl-3,4-dihydroisoquinoline-2(1H)-sulfonamide | | Descriptor: | (1R)-6,7-dimethoxy-1-methyl-3,4-dihydroisoquinoline-2(1H)-sulfonamide, (1S)-6,7-dimethoxy-1-methyl-3,4-dihydroisoquinoline-2(1H)-sulfonamide, ACETATE ION, ... | | Authors: | Mader, P, Brynda, J, Gitto, R, Agnello, S, Ferro, S, De Luca, L, Vullo, D, Supuran, C.T, Chimirri, A. | | Deposit date: | 2010-11-22 | | Release date: | 2011-04-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural basis for the interaction between carbonic anhydrase and 1,2,3,4-tetrahydroisoquinolin-2-ylsulfonamides.
J.Med.Chem., 54, 2011
|
|
6G02
 
 | | Complex of neuraminidase from H1N1 influenza virus with tamiphosphor omega-azidohexyl ester | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-[[(3~{R},4~{R},5~{S})-4-acetamido-5-azanyl-3-pentan-3-yloxy-cyclohexen-1-yl]-oxidanyl-phosphoryl]oxyhexylimino-azanylidene-azanium, ... | | Authors: | Pachl, P, Pokorna, J. | | Deposit date: | 2018-03-15 | | Release date: | 2019-01-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | DNA-linked inhibitor antibody assay (DIANA) as a new method for screening influenza neuraminidase inhibitors.
Biochem. J., 475, 2018
|
|
6HP0
 
 | | Complex of Neuraminidase from H1N1 Influenza Virus in Complex with Oseltamivir Triazol Derivative | | Descriptor: | (3~{R},4~{R},5~{S})-4-acetamido-5-[4-(hydroxymethyl)-1,2,3-triazol-1-yl]-3-pentan-3-yloxy-cyclohexene-1-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pachl, P, Pokorna, J. | | Deposit date: | 2018-09-19 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Investigation of flexibility of neuraminidase 150-loop using tamiflu derivatives in influenza A viruses H1N1 and H5N1.
Bioorg.Med.Chem., 27, 2019
|
|
2HKH
 
 | | Crystal structure of the Fab M75 | | Descriptor: | GLYCEROL, Immunoglobulin Heavy chain Fab fragment, Immunoglobulin Light chain Fab fragment | | Authors: | Kral, V, Mader, P, Stouracova, R, Fabry, M, Sedlacek, J, Brynda, J. | | Deposit date: | 2006-07-04 | | Release date: | 2007-11-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Stabilization of antibody structure upon association to a human carbonic anhydrase IX epitope studied by X-ray crystallography, microcalorimetry, and molecular dynamics simulations.
Proteins, 71, 2008
|
|
2HKF
 
 | | Crystal structure of the Complex Fab M75- Peptide | | Descriptor: | Carbonic anhydrase 9, Immunoglobulin Heavy chain Fab fragment, Immunoglobulin Light chain Fab fragment | | Authors: | Kral, V, Mader, P, Stouracova, R, Fabry, M, Horejsi, M, Brynda, J. | | Deposit date: | 2006-07-04 | | Release date: | 2007-11-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Stabilization of antibody structure upon association to a human carbonic anhydrase IX epitope studied by X-ray crystallography, microcalorimetry, and molecular dynamics simulations.
Proteins, 71, 2008
|
|
3CCK
 
 | | Human CD69 | | Descriptor: | CHLORIDE ION, Early activation antigen CD69 | | Authors: | Brynda, J, Vanek, O, Rezacova, P. | | Deposit date: | 2008-02-26 | | Release date: | 2008-11-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Soluble recombinant CD69 receptors optimized to have an exceptional physical and chemical stability display prolonged circulation and remain intact in the blood of mice
Febs J., 275, 2008
|
|
2Z54
 
 | | The Influence of I47A Mutation on Reduced Susceptibility to the Protease Inhibitor Lopinavir | | Descriptor: | BETA-MERCAPTOETHANOL, HIV-1 Protease, N-{1-BENZYL-4-[2-(2,6-DIMETHYL-PHENOXY)-ACETYLAMINO]-3-HYDROXY-5-PHENYL-PENTYL}-3-METHYL-2-(2-OXO-TETRAHYDRO-PYRIMIDIN-1-YL)-BUTYRAMIDE | | Authors: | Brynda, J, Klara, S, Kozisek, M, Lepsik, M, Machala, L, Konvalinka, J. | | Deposit date: | 2007-06-28 | | Release date: | 2008-07-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Enzymatic and structural analysis of the I47A mutation contributing to the reduced susceptibility to HIV protease inhibitor lopinavir.
Protein Sci., 17, 2008
|
|
8A2H
 
 | | human STING in complex with 2',3'-cyclic-GMP-7-deazaphenyl-AMP | | Descriptor: | 2-azanyl-9-[(1~{R},6~{R},8~{R},9~{R},10~{S},15~{R},17~{R},18~{R})-8-(4-azanyl-5-phenyl-pyrrolo[2,3-d]pyrimidin-7-yl)-3,9,12,18-tetrakis(oxidanyl)-3,12-bis(oxidanylidene)-2,4,7,11,13,16-hexaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.2.1.0^{6,10}]octadecan-17-yl]-1~{H}-purin-6-one, Stimulator of interferon genes protein | | Authors: | Smola, M, Vavrina, Z, Boura, E, Brynda, J. | | Deposit date: | 2022-06-03 | | Release date: | 2022-10-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Design, Synthesis, and Biochemical and Biological Evaluation of Novel 7-Deazapurine Cyclic Dinucleotide Analogues as STING Receptor Agonists.
J.Med.Chem., 65, 2022
|
|
2QHC
 
 | | The Influence of I47A Mutation on Reduced Susceptibility to the Protease Inhibitor Lopinavir | | Descriptor: | BETA-MERCAPTOETHANOL, N-{1-BENZYL-4-[2-(2,6-DIMETHYL-PHENOXY)-ACETYLAMINO]-3-HYDROXY-5-PHENYL-PENTYL}-3-METHYL-2-(2-OXO-TETRAHYDRO-PYRIMIDIN-1-YL)-BUTYRAMIDE, PROTEASE RETROPEPSIN | | Authors: | Brynda, J, Saskova, K.G, Kozisek, M, Lepsik, M, Machala, L, Konvalinka, J. | | Deposit date: | 2007-07-02 | | Release date: | 2008-07-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Enzymatic and structural analysis of the I47A mutation contributing to the reduced susceptibility to HIV protease inhibitor lopinavir.
Protein Sci., 17, 2008
|
|
