1F04
 
 | | SOLUTION STRUCTURE OF OXIDIZED BOVINE MICROSOMAL CYTOCHROME B5 MUTANT (E44A, E48A, E56A, D60A) AND ITS INTERACTION WITH CYTOCHROME C | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wu, Y.B, Lu, J, Qian, C.M, Tang, W.X, Li, E.C, Wang, J.F, Wang, Y.H, Wang, W.H, Lu, J.X, Xie, Y, Huang, Z.X. | | Deposit date: | 2000-05-14 | | Release date: | 2000-06-21 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Solution structure of cytochrome b(5) mutant (E44/48/56A/D60A) and its interaction with cytochrome c.
Eur.J.Biochem., 268, 2001
|
|
7XOE
 
 | | Cryo-EM structure of S glycoprotein encoded by the Covid-19 mRNA vaccine candidate RQ3013 (Prefusion state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,peptide | | Authors: | Wu, Z, Yu, Z, Tan, S, Lu, J, Lu, G, Lin, J. | | Deposit date: | 2022-05-01 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Preclinical evaluation of RQ3013, a broad-spectrum mRNA vaccine against SARS-CoV-2 variants.
Sci Bull (Beijing), 68, 2023
|
|
7XOG
 
 | | Cryo-EM structure of S glycoprotein encoded by the Covid-19 mRNA vaccine candidate RQ3013 (Postfusion state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,peptide, ... | | Authors: | Wu, Z, Yu, Z, Tan, S, Lu, J, Lu, G, Lin, J. | | Deposit date: | 2022-05-01 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Preclinical evaluation of RQ3013, a broad-spectrum mRNA vaccine against SARS-CoV-2 variants.
Sci Bull (Beijing), 68, 2023
|
|
2A20
 
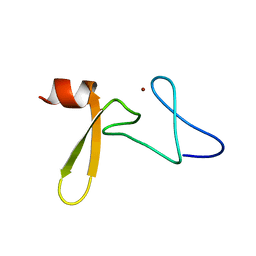 | | Solution structure of Rim2 Zinc Finger Domain | | Descriptor: | Regulating synaptic membrane exocytosis protein 2, ZINC ION | | Authors: | Dulubova, I, Lou, X, Lu, J, Huryeva, I, Alam, A, Schneggenburger, R, Sudhof, T.C, Rizo, J. | | Deposit date: | 2005-06-21 | | Release date: | 2005-08-16 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | A Munc13/RIM/Rab3 tripartite complex: from priming to plasticity?
Embo J., 24, 2005
|
|
8T1U
 
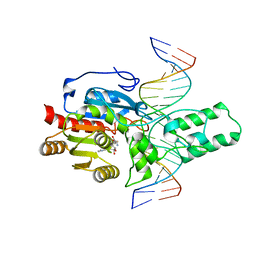 | | Crystal structure of the DRM2-CTA DNA complex | | Descriptor: | DNA (5'-D(P*AP*TP*TP*AP*TP*TP*AP*AP*TP*(C49)P*TP*AP*AP*AP*TP*TP*TP*A)-3'), DNA (5'-D(P*TP*AP*AP*AP*TP*TP*TP*AP*GP*AP*TP*TP*AP*AP*TP*AP*AP*T)-3'), DNA (cytosine-5)-methyltransferase DRM2, ... | | Authors: | Chen, J, Lu, J, Song, J. | | Deposit date: | 2023-06-03 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | DNA conformational dynamics in the context-dependent non-CG CHH methylation by plant methyltransferase DRM2.
J.Biol.Chem., 299, 2023
|
|
2WZZ
 
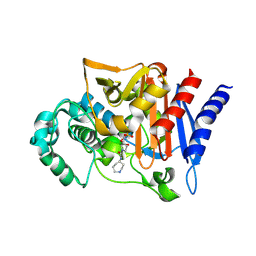 | |
2WZX
 
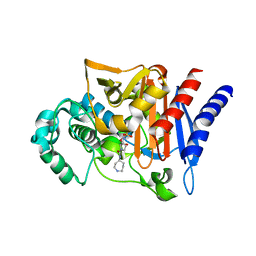 | |
3IL5
 
 | | Structure of E. faecalis FabH in complex with 2-({4-bromo-3-[(diethylamino)sulfonyl]benzoyl}amino)benzoic acid | | Descriptor: | 2-({[4-bromo-3-(diethylsulfamoyl)phenyl]carbonyl}amino)benzoic acid, 3-oxoacyl-[acyl-carrier-protein] synthase 3 | | Authors: | Gajiwala, K.S, Margosiak, S, Lu, J, Cortez, J, Su, Y, Nie, Z, Appelt, K. | | Deposit date: | 2009-08-06 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of bacterial FabH suggest a molecular basis for the substrate specificity of the enzyme.
Febs Lett., 583, 2009
|
|
3IL6
 
 | | Structure of E. faecalis FabH in complex with 2-({4-[(3R,5S)-3,5-dimethylpiperidin-1-yl]-3-phenoxybenzoyl}amino)benzoic acid | | Descriptor: | 2-[({4-[(3R,5S)-3,5-dimethylpiperidin-1-yl]-3-phenoxyphenyl}carbonyl)amino]benzoic acid, 3-oxoacyl-[acyl-carrier-protein] synthase 3, SULFATE ION | | Authors: | Gajiwala, K.S, Margosiak, S, Lu, J, Cortez, J, Su, Y, Nie, Z, Appelt, K. | | Deposit date: | 2009-08-06 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of bacterial FabH suggest a molecular basis for the substrate specificity of the enzyme.
Febs Lett., 583, 2009
|
|
3IL9
 
 | | Structure of E. coli FabH | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 3 | | Authors: | Gajiwala, K.S, Margosiak, S, Lu, J, Cortez, J, Su, Y, Nie, Z, Appelt, K. | | Deposit date: | 2009-08-06 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of bacterial FabH suggest a molecular basis for the substrate specificity of the enzyme.
Febs Lett., 583, 2009
|
|
3IL7
 
 | | Crystal structure of S. aureus FabH | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 3 | | Authors: | Gajiwala, K.S, Margosiak, S, Lu, J, Cortez, J, Su, Y, Nie, Z, Appelt, K. | | Deposit date: | 2009-08-06 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of bacterial FabH suggest a molecular basis for the substrate specificity of the enzyme.
Febs Lett., 583, 2009
|
|
3IL3
 
 | | Structure of Haemophilus influenzae FabH | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 3 | | Authors: | Gajiwala, K.S, Margosiak, S, Lu, J, Cortez, J, Su, Y, Nie, Z, Appelt, K. | | Deposit date: | 2009-08-06 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of bacterial FabH suggest a molecular basis for the substrate specificity of the enzyme.
Febs Lett., 583, 2009
|
|
3IL4
 
 | | Structure of E. faecalis FabH in complex with acetyl CoA | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 3, ACETYL COENZYME *A | | Authors: | Gajiwala, K.S, Margosiak, S, Lu, J, Cortez, J, Su, Y, Nie, Z, Appelt, K. | | Deposit date: | 2009-08-06 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of bacterial FabH suggest a molecular basis for the substrate specificity of the enzyme.
Febs Lett., 583, 2009
|
|
6NOV
 
 | | A Fab derived from ixekizumab | | Descriptor: | DODECAETHYLENE GLYCOL, Fab Heavy Chain, Fab Light Chain | | Authors: | Durbin, J.D, Clawson, D.K, Lu, F, Tian, Y, Lu, J, Schmitt, M, Atwell, S. | | Deposit date: | 2019-01-16 | | Release date: | 2019-06-19 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Development of tibulizumab, a tetravalent bispecific antibody targeting BAFF and IL-17A for the treatment of autoimmune disease.
Mabs, 11, 2019
|
|
7MBI
 
 | | Structure of SARS-CoV2 3CL protease covalently bound to peptidomimetic inhibitor | | Descriptor: | 2,4,6-trimethylpyridine-3-carboxylic acid, 3C-like proteinase, 4-methoxy-N-[(2S)-4-methyl-1-oxo-1-({(2S)-3-oxo-1-[(3S)-2-oxopiperidin-3-yl]butan-2-yl}amino)pentan-2-yl]-1H-indole-2-carboxamide | | Authors: | Khan, M.B, Lu, J, Young, H.S, Lemieux, M.J. | | Deposit date: | 2021-03-31 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Peptidomimetic alpha-Acyloxymethylketone Warheads with Six-Membered Lactam P1 Glutamine Mimic: SARS-CoV-2 3CL Protease Inhibition, Coronavirus Antiviral Activity, and in Vitro Biological Stability.
J.Med.Chem., 65, 2022
|
|
3BE9
 
 | | Structure-based design and synthesis of novel macrocyclic pyrazolo[1,5-a] [1,3,5]triazine compounds as potent inhibitors of protein kinase CK2 and their anticancer activities | | Descriptor: | 19-(cyclopropylamino)-4,6,7,15-tetrahydro-5H-16,1-(azenometheno)-10,14-(metheno)pyrazolo[4,3-o][1,3,9]triazacyclohexadecin-8(9H)-one, Casein kinase II subunit alpha | | Authors: | Nie, Z, Perretta, C, Erickson, P, Margosiak, S, Lu, J, Averill, A, Almassy, R, Chu, S. | | Deposit date: | 2007-11-16 | | Release date: | 2008-11-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design and synthesis of novel macrocyclic pyrazolo[1,5-a] [1,3,5]triazine compounds as potent inhibitors of protein kinase CK2 and their anticancer activities.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
6VIL
 
 | |
8T12
 
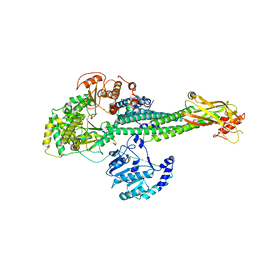 | |
8T13
 
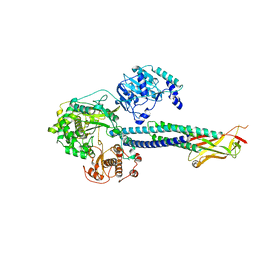 | |
2H9C
 
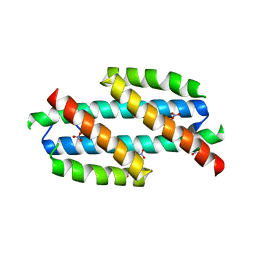 | |
4ZQD
 
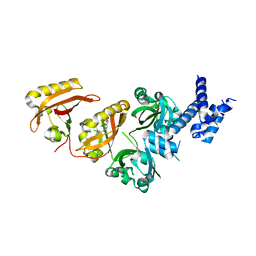 | | Crystal Structure of the Heterodimeric HIF-2a:ARNT Complex with the Benzoxadiazole Antagonist 0X3 | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1, N-(3-chloro-5-fluorophenyl)-4-nitro-2,1,3-benzoxadiazol-5-amine | | Authors: | Wu, D, Potluri, N, Lu, J, Kim, Y, Rastinejad, F. | | Deposit date: | 2015-05-09 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Structural integration in hypoxia-inducible factors.
Nature, 524, 2015
|
|
4ZPR
 
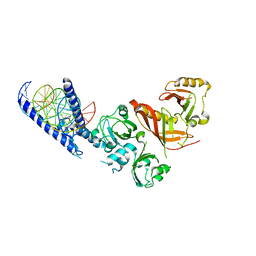 | | Crystal Structure of the Heterodimeric HIF-1a:ARNT Complex with HRE DNA | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, DNA (5'-D(*CP*AP*CP*GP*AP*CP*CP*CP*GP*CP*AP*CP*GP*TP*AP*CP*GP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*GP*CP*TP*GP*CP*GP*TP*AP*CP*GP*TP*GP*CP*GP*GP*GP*TP*CP*GP*T)-3'), ... | | Authors: | Wu, D, Potluri, N, Lu, J, Kim, Y, Rastinejad, F. | | Deposit date: | 2015-05-08 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.902 Å) | | Cite: | Structural integration in hypoxia-inducible factors.
Nature, 524, 2015
|
|
4ZPK
 
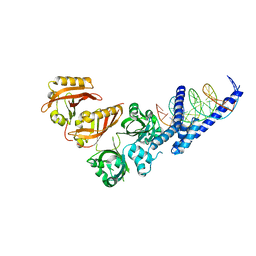 | | Crystal Structure of the Heterodimeric HIF-2a:ARNT Complex with HRE DNA | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, DNA (5'-D(*CP*AP*CP*GP*AP*CP*CP*CP*GP*CP*AP*CP*GP*TP*AP*CP*GP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*GP*CP*TP*GP*CP*GP*TP*AP*CP*GP*TP*GP*CP*GP*GP*GP*TP*CP*GP*T)-3'), ... | | Authors: | Wu, D, Potluri, N, Lu, J, Kim, Y, Rastinejad, F. | | Deposit date: | 2015-05-08 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural integration in hypoxia-inducible factors.
Nature, 524, 2015
|
|
4ZP4
 
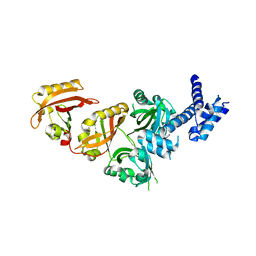 | | Crystal Structure of the Heterodimeric HIF-2a:ARNT Complex | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Wu, D, Potluri, N, Lu, J, Kim, Y, Rastinejad, F. | | Deposit date: | 2015-05-07 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.355 Å) | | Cite: | Structural integration in hypoxia-inducible factors.
Nature, 524, 2015
|
|
4ZPH
 
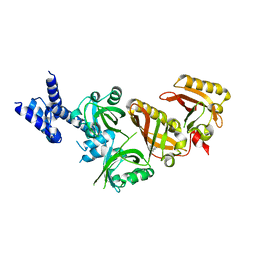 | | Crystal Structure of the Heterodimeric HIF-2a:ARNT Complex with Proflavine | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1, PROFLAVIN | | Authors: | Wu, D, Potluri, N, Lu, J, Kim, Y, Rastinejad, F. | | Deposit date: | 2015-05-07 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural integration in hypoxia-inducible factors.
Nature, 524, 2015
|
|
