6KVI
 
 | | Crystal structure of UDP-SrUGT76G1 | | 分子名称: | UDP-glycosyltransferase 76G1, URIDINE-5'-DIPHOSPHATE | | 著者 | Li, J.X, Liu, Z.F, Wang, Y, Zhang, P. | | 登録日 | 2019-09-04 | | 公開日 | 2019-11-20 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.598 Å) | | 主引用文献 | Structural Insights into the Catalytic Mechanism of a Plant Diterpene Glycosyltransferase SrUGT76G1.
Plant Commun., 1, 2020
|
|
6KVJ
 
 | | Crystal structure of UDPX-SrUGT76G1 | | 分子名称: | UDP-glycosyltransferase 76G1, URIDINE-5'-DIPHOSPHATE-XYLOPYRANOSE | | 著者 | Li, J.X, Liu, Z.F, Wang, Y, Zhang, P. | | 登録日 | 2019-09-04 | | 公開日 | 2019-11-20 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.499 Å) | | 主引用文献 | Structural Insights into the Catalytic Mechanism of a Plant Diterpene Glycosyltransferase SrUGT76G1.
Plant Commun., 1, 2020
|
|
6KVK
 
 | | Crystal structure of UDP-Sm-SrUGT76G1 | | 分子名称: | Steviolmonoside, UDP-glycosyltransferase 76G1, URIDINE-5'-DIPHOSPHATE | | 著者 | Li, J.X, Liu, Z.F, Wang, Y, Zhang, P. | | 登録日 | 2019-09-04 | | 公開日 | 2019-11-20 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.397 Å) | | 主引用文献 | Structural Insights into the Catalytic Mechanism of a Plant Diterpene Glycosyltransferase SrUGT76G1.
Plant Commun., 1, 2020
|
|
7V26
 
 | | XG005-bound SARS-CoV-2 S | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, XG005 Heavy chain, ... | | 著者 | Zhan, W.Q, Zhang, X, Sun, L, Chen, Z.G. | | 登録日 | 2021-08-07 | | 公開日 | 2021-10-20 | | 最終更新日 | 2022-07-06 | | 実験手法 | ELECTRON MICROSCOPY (3.85 Å) | | 主引用文献 | An ultrapotent pan-beta-coronavirus lineage B ( beta-CoV-B) neutralizing antibody locks the receptor-binding domain in closed conformation by targeting its conserved epitope.
Protein Cell, 13, 2022
|
|
7V2A
 
 | | SARS-CoV-2 Spike trimer in complex with XG014 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, The heavy chain of XG014, ... | | 著者 | Wang, K, Wang, X, Pan, L. | | 登録日 | 2021-08-07 | | 公開日 | 2021-10-20 | | 最終更新日 | 2022-07-06 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | An ultrapotent pan-beta-coronavirus lineage B ( beta-CoV-B) neutralizing antibody locks the receptor-binding domain in closed conformation by targeting its conserved epitope.
Protein Cell, 13, 2022
|
|
7WCV
 
 | | Co-crystal structure of FTO bound to 6e | | 分子名称: | 2-OXOGLUTARIC ACID, 2-[[2,6-bis(chloranyl)-4-pyridin-4-yl-phenyl]amino]benzoic acid, Alpha-ketoglutarate-dependent dioxygenase FTO, ... | | 著者 | Yang, C.-G, Gan, J.H. | | 登録日 | 2021-12-20 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure-Activity Relationships and Antileukemia Effects of the Tricyclic Benzoic Acid FTO Inhibitors.
J.Med.Chem., 65, 2022
|
|
7UIE
 
 | | Crystal structure of HcE-JLE-G6 | | 分子名称: | Botulinum neurotoxin E heavy chain, JLE-G6 | | 著者 | Jin, R, Lam, K. | | 登録日 | 2022-03-29 | | 公開日 | 2023-04-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.23 Å) | | 主引用文献 | Structural basis for botulinum neurotoxin E recognition of synaptic vesicle protein 2.
Nat Commun, 14, 2023
|
|
8X1N
 
 | | Cryo-EM structure of human alpha-fetoprotein | | 分子名称: | Alpha-fetoprotein, PALMITIC ACID, ZINC ION, ... | | 著者 | Liu, Z.M, Li, M.S, Wu, C, Liu, K. | | 登録日 | 2023-11-08 | | 公開日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.31 Å) | | 主引用文献 | Structural characteristics of alpha-fetoprotein, including N-glycosylation, metal ion and fatty acid binding sites.
Commun Biol, 7, 2024
|
|
4Y5U
 
 | | Transcription factor | | 分子名称: | NICKEL (II) ION, Signal transducer and activator of transcription 6 | | 著者 | Li, J, Niu, F, Ouyang, S, Liu, Z. | | 登録日 | 2015-02-12 | | 公開日 | 2016-02-17 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.708 Å) | | 主引用文献 | Structural basis for DNA recognition by STAT6
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4Y5W
 
 | | Transcription factor-DNA complex | | 分子名称: | DNA (5'-D(P*AP*TP*GP*GP*AP*TP*TP*TP*CP*CP*TP*AP*GP*GP*AP*AP*GP*AP*CP*AP*A)-3'), DNA (5'-D(P*TP*TP*GP*TP*CP*TP*TP*CP*CP*TP*AP*GP*GP*AP*AP*AP*TP*CP*CP*AP*T)-3'), Signal transducer and activator of transcription 6 | | 著者 | Li, J, Niu, F, Ouyang, S, Liu, Z. | | 登録日 | 2015-02-12 | | 公開日 | 2016-02-17 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (3.104 Å) | | 主引用文献 | Structural basis for DNA recognition by STAT6
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
8W5K
 
 | | Cryo-EM structure of the yeast TOM core complex crosslinked by BS3 (from TOM-TIM23 complex) | | 分子名称: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradecanoyloxy)propyl tetradecanoate, Mitochondrial import receptor subunit TOM22, Mitochondrial import receptor subunit TOM40, ... | | 著者 | Wang, Q, Guan, Z.Y, Zhuang, J.J, Huang, R, Yin, P. | | 登録日 | 2023-08-27 | | 公開日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | The architecture of substrate-engaged TOM-TIM23 supercomplex reveals preprotein proximity sites for mitochondrial protein translocation.
Cell Discov, 10, 2024
|
|
8W5J
 
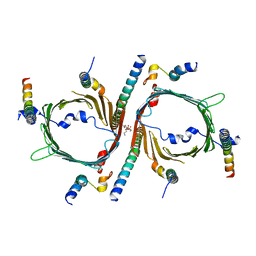 | | Cryo-EM structure of the yeast TOM core complex (from TOM-TIM23 complex) | | 分子名称: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradecanoyloxy)propyl tetradecanoate, Mitochondrial import receptor subunit TOM22, Mitochondrial import receptor subunit TOM40, ... | | 著者 | Wang, Q, Guan, Z.Y, Zhuang, J.J, Huang, R, Yin, P. | | 登録日 | 2023-08-27 | | 公開日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | The architecture of substrate-engaged TOM-TIM23 supercomplex reveals preprotein proximity sites for mitochondrial protein translocation.
Cell Discov, 10, 2024
|
|
7BZF
 
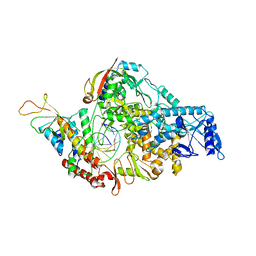 | | COVID-19 RNA-dependent RNA polymerase post-translocated catalytic complex | | 分子名称: | Non-structural protein 7, Non-structural protein 8, RNA (31-MER), ... | | 著者 | Wang, Q, Gao, Y, Ji, W, Mu, A, Rao, Z. | | 登録日 | 2020-04-27 | | 公開日 | 2020-06-03 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.26 Å) | | 主引用文献 | Structural Basis for RNA Replication by the SARS-CoV-2 Polymerase.
Cell, 182, 2020
|
|
7C2K
 
 | | COVID-19 RNA-dependent RNA polymerase pre-translocated catalytic complex | | 分子名称: | Non-structural protein 7, Non-structural protein 8, RNA (29-MER), ... | | 著者 | Wang, Q, Gao, Y, Ji, W, Mu, A, Rao, Z. | | 登録日 | 2020-05-07 | | 公開日 | 2020-06-03 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (2.93 Å) | | 主引用文献 | Structural Basis for RNA Replication by the SARS-CoV-2 Polymerase.
Cell, 182, 2020
|
|
7BTF
 
 | | SARS-CoV-2 RNA-dependent RNA polymerase in complex with cofactors in reduced condition | | 分子名称: | Non-structural protein 7, Non-structural protein 8, RNA-directed RNA polymerase, ... | | 著者 | Gao, Y, Yan, L, Huang, Y, Liu, F, Cao, L, Wang, T, Wang, Q, Lou, Z, Rao, Z. | | 登録日 | 2020-04-01 | | 公開日 | 2020-04-08 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (2.95 Å) | | 主引用文献 | Structure of the RNA-dependent RNA polymerase from COVID-19 virus.
Science, 368, 2020
|
|
8GCM
 
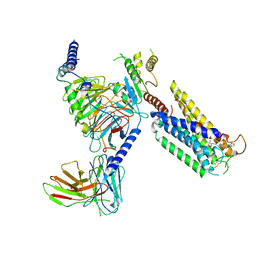 | | Cryo-EM Structure of the Prostaglandin E Receptor EP4 Coupled to G Protein | | 分子名称: | (5S)-5-[(3R)-4,4-difluoro-3-hydroxy-4-phenylbutyl]-1-[6-(1H-tetrazol-5-yl)hexyl]pyrrolidin-2-one, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Huang, S.M, Xiong, M.Y, Liu, L, Mu, J, Sheng, C, Sun, J. | | 登録日 | 2023-03-02 | | 公開日 | 2024-01-03 | | 最終更新日 | 2024-01-10 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Single hormone or synthetic agonist induces G s /G i coupling selectivity of EP receptors via distinct binding modes and propagating paths.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
1RYG
 
 | |
1RYV
 
 | |
4EBZ
 
 | | Crystal structure of the ectodomain of a receptor like kinase | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitin elicitor receptor kinase 1, ... | | 著者 | Chai, J, Liu, T, Han, Z, She, J, Wang, J. | | 登録日 | 2012-03-26 | | 公開日 | 2012-06-27 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.792 Å) | | 主引用文献 | Chitin-induced dimerization activates a plant immune receptor.
Science, 336, 2012
|
|
4EBY
 
 | | Crystal structure of the ectodomain of a receptor like kinase | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitin elicitor receptor kinase 1, ... | | 著者 | Chai, J, Liu, T, Han, Z, She, J, Wang, J. | | 登録日 | 2012-03-25 | | 公開日 | 2012-06-27 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Chitin-induced dimerization activates a plant immune receptor.
Science, 336, 2012
|
|
7CJM
 
 | | SARS CoV-2 PLpro in complex with GRL0617 | | 分子名称: | 5-amino-2-methyl-N-[(1R)-1-naphthalen-1-ylethyl]benzamide, Non-structural protein 3, ZINC ION | | 著者 | Fu, Z, Huang, H. | | 登録日 | 2020-07-11 | | 公開日 | 2020-09-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | The complex structure of GRL0617 and SARS-CoV-2 PLpro reveals a hot spot for antiviral drug discovery.
Nat Commun, 12, 2021
|
|
7YTO
 
 | |
7C1D
 
 | |
5L0Q
 
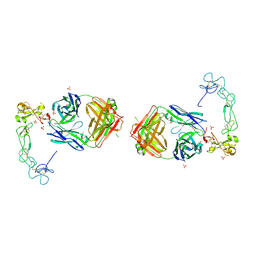 | | Crystal structure of the complex between ADAM10 D+C domain and a conformation specific mAb 8C7. | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Disintegrin and metalloproteinase domain-containing protein 10, MAGNESIUM ION, ... | | 著者 | Xu, K, Saha, N, Nikolov, D.B. | | 登録日 | 2016-07-28 | | 公開日 | 2016-11-09 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.759 Å) | | 主引用文献 | An activated form of ADAM10 is tumor selective and regulates cancer stem-like cells and tumor growth.
J.Exp.Med., 213, 2016
|
|
6ILM
 
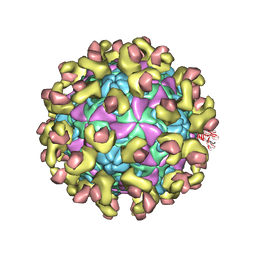 | | Cryo-EM structure of Echovirus 6 complexed with its uncoating receptor FcRn at PH 7.4 | | 分子名称: | Beta-2-microglobulin, Capsid protein VP1, Capsid protein VP2, ... | | 著者 | Gao, G.F, Liu, S, Zhao, X, Peng, R. | | 登録日 | 2018-10-19 | | 公開日 | 2019-05-15 | | 最終更新日 | 2019-11-06 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Human Neonatal Fc Receptor Is the Cellular Uncoating Receptor for Enterovirus B.
Cell, 177, 2019
|
|
