3U57
 
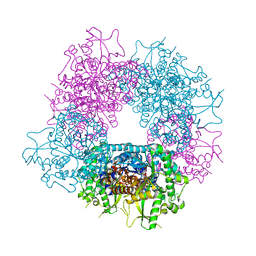 | | Structures of Alkaloid Biosynthetic Glucosidases Decode Substrate Specificity | | Descriptor: | (2beta,7beta,16S,17R,19E,21beta)-21-(beta-D-glucopyranosyloxy)-2,7-dihydro-7,17-cyclosarpagan-17-yl acetate, CHLORIDE ION, Raucaffricine-O-beta-D-glucosidase | | Authors: | Xia, L, Ruppert, M, Wang, M, Panjikar, S, Lin, H, Rajendran, C, Barleben, L, Stoeckigt, J. | | Deposit date: | 2011-10-11 | | Release date: | 2011-11-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structures of alkaloid biosynthetic glucosidases decode substrate specificity.
Acs Chem.Biol., 7, 2012
|
|
3U5Y
 
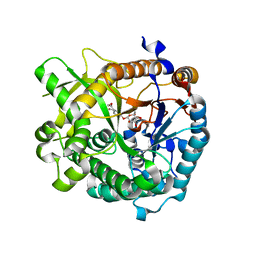 | | Structures of Alkaloid Biosynthetic Glucosidases Decode Substrate Specificity | | Descriptor: | CHLORIDE ION, Raucaffricine-O-beta-D-glucosidase, Secologanin | | Authors: | Xia, L, Ruppert, M, Wang, M, Panjikar, S, Lin, H, Rajendran, C, Barleben, L, Stoeckigt, J. | | Deposit date: | 2011-10-11 | | Release date: | 2011-11-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of alkaloid biosynthetic glucosidases decode substrate specificity.
Acs Chem.Biol., 7, 2012
|
|
2KXQ
 
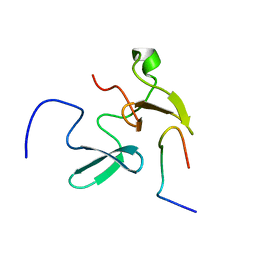 | | Solution Structure of Smurf2 WW2 and WW3 bound to Smad7 PY motif containing peptide | | Descriptor: | E3 ubiquitin-protein ligase SMURF2, Smad7 PY motif containing peptide | | Authors: | Chong, A, Lin, H, Wrana, J, Forman-Kay, J.D. | | Deposit date: | 2010-05-11 | | Release date: | 2010-10-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Coupling of tandem Smad ubiquitination regulatory factor (Smurf) WW domains modulates target specificity.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6LU5
 
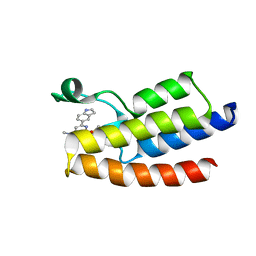 | | Crystal structure of BPTF-BRD with ligand DCBPin5 bound | | Descriptor: | 6-(1H-indol-5-yl)-N-methyl-2-methylsulfonyl-pyrimidin-4-amine, Nucleosome-remodeling factor subunit BPTF | | Authors: | Lu, T, Lu, H.B, Wang, J, Lin, H, Lu, W, Luo, C. | | Deposit date: | 2020-01-25 | | Release date: | 2021-04-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.86527729 Å) | | Cite: | Discovery and Optimization of Small-Molecule Inhibitors for the BPTF Bromodomains Proteins
To Be Published
|
|
6LU6
 
 | | Crystal structure of BPTF-BRD with ligand DCBPin5-2 bound | | Descriptor: | 6-[1-[3-(dimethylamino)propyl]indol-5-yl]-2-methylsulfonyl-N-propyl-pyrimidin-4-amine, Nucleosome-remodeling factor subunit BPTF | | Authors: | Lu, T, Lu, H.B, Wang, J, Lin, H, Lu, W, Luo, C. | | Deposit date: | 2020-01-26 | | Release date: | 2021-04-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.970063 Å) | | Cite: | Discovery and Optimization of Small-Molecule Inhibitors for the BPTF Bromodomains Proteins
To Be Published
|
|
6M61
 
 | | Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) with inhibitor heptelidic acid | | Descriptor: | (5aS,6R,9S,9aS)-9-methyl-9-oxidanyl-1-oxidanylidene-6-propan-2-yl-3,5a,6,7,8,9a-hexahydro-2-benzoxepine-4-carboxylic acid, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yan, Y, Zang, X, Cooper, S.J, Lin, H, Zhou, J, Tang, Y. | | Deposit date: | 2020-03-12 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.82449543 Å) | | Cite: | Biosynthesis of the fungal glyceraldehyde-3-phosphate dehydrogenase inhibitor heptelidic acid and mechanism of self-resistance
Chem Sci, 11, 2020
|
|
6M5X
 
 | | A fungal glyceraldehyde-3-phosphate dehydrogenase with self-resistance to inhibitor heptelidic acid | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Yan, Y, Zang, X, Cooper, S.J, Lin, H, Zhou, J, Tang, Y. | | Deposit date: | 2020-03-12 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05990934 Å) | | Cite: | Biosynthesis of the fungal glyceraldehyde-3-phosphate dehydrogenase inhibitor heptelidic acid and mechanism of self-resistance
Chem Sci, 11, 2020
|
|
7F5D
 
 | | Crystal structure of BPTF-BRD with ligand DC-BPi-03 bound | | Descriptor: | 6-(1H-indol-5-yl)-N-methyl-2-methylsulfonyl-pyrimidin-4-amine, Nucleosome-remodeling factor subunit BPTF | | Authors: | Lu, T, Lu, H.B, Wang, J, Lin, H, Lu, W, Luo, C. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.57150865 Å) | | Cite: | Discovery and Optimization of Small-Molecule Inhibitors for the BPTF Bromodomains Proteins
To Be Published
|
|
7F5E
 
 | | Crystal structure of BPTF-BRD with ligand DC-BPi-11 bound | | Descriptor: | N,N-dimethyl-3-[5-(2-methylsulfonyl-7H-pyrrolo[2,3-d]pyrimidin-4-yl)indol-1-yl]propan-1-amine, Nucleosome-remodeling factor subunit BPTF | | Authors: | Lu, T, Lu, H.B, Wang, J, Lin, H, Lu, W, Luo, C. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.20017123 Å) | | Cite: | Discovery and Optimization of Small-Molecule Inhibitors for the BPTF Bromodomains Proteins
To Be Published
|
|
7F5C
 
 | | Crystal structure of BPTF-BRD with ligand DC-BPi-07 bound | | Descriptor: | 6-[1-[3-(dimethylamino)propyl]indol-5-yl]-2-methylsulfonyl-N-propyl-pyrimidin-4-amine, Nucleosome-remodeling factor subunit BPTF | | Authors: | Lu, T, Lu, H.B, Wang, J, Lin, H, Lu, W, Luo, C. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65004492 Å) | | Cite: | Discovery and Optimization of Small-Molecule Inhibitors for the BPTF Bromodomains Proteins
To Be Published
|
|
6BXL
 
 | | Crystal structure of Pyrococcus horikoshii Dph2 with 4Fe-4S cluster and SAM | | Descriptor: | 2-(3-amino-3-carboxypropyl)histidine synthase, IRON/SULFUR CLUSTER, S-ADENOSYLMETHIONINE | | Authors: | Torelli, A.T, Fenwick, M.K, Zhang, Y, Dong, M, Kathiresan, V, Carantoa, J.D, Dzikovski, B, Lancaster, K.M, Freed, J.H, Hoffman, B.M, Lin, H, Ealick, S.E. | | Deposit date: | 2017-12-18 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Organometallic and radical intermediates reveal mechanism of diphthamide biosynthesis.
Science, 359, 2018
|
|
6BXM
 
 | | Crystal structure of Candidatus Methanoperedens nitroreducens Dph2 with 4Fe-4S cluster and SAM/cleaved SAM | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, ALPHA-AMINOBUTYRIC ACID, Diphthamide biosynthesis enzyme Dph2, ... | | Authors: | Fenwick, M.K, Torelli, A.T, Zhang, Y, Dong, M, Kathiresan, V, Carantoa, J.D, Dzikovski, B, Lancaster, K.M, Freed, J.H, Hoffman, B.M, Lin, H, Ealick, S.E. | | Deposit date: | 2017-12-18 | | Release date: | 2018-04-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.252 Å) | | Cite: | Organometallic and radical intermediates reveal mechanism of diphthamide biosynthesis.
Science, 359, 2018
|
|
6BXN
 
 | | Crystal structure of Candidatus Methanoperedens nitroreducens Dph2 with 4Fe-4S cluster and SAM | | Descriptor: | Diphthamide biosynthesis enzyme Dph2, IRON/SULFUR CLUSTER, S-ADENOSYLMETHIONINE | | Authors: | Fenwick, M.K, Torelli, A.T, Zhang, Y, Dong, M, Kathiresan, V, Carantoa, J.D, Dzikovski, B, Lancaster, K.M, Freed, J.H, Hoffman, B.M, Lin, H, Ealick, S.E. | | Deposit date: | 2017-12-18 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.079 Å) | | Cite: | Organometallic and radical intermediates reveal mechanism of diphthamide biosynthesis.
Science, 359, 2018
|
|
6BXK
 
 | | Crystal structure of Pyrococcus horikoshii Dph2 with 4Fe-4S cluster and MTA | | Descriptor: | 2-(3-amino-3-carboxypropyl)histidine synthase, 5'-DEOXY-5'-METHYLTHIOADENOSINE, IRON/SULFUR CLUSTER, ... | | Authors: | Torelli, A.T, Fenwick, M.K, Zhang, Y, Dong, M, Kathiresan, V, Carantoa, J.D, Dzikovski, B, Lancaster, K.M, Freed, J.H, Hoffman, B.M, Lin, H, Ealick, S.E. | | Deposit date: | 2017-12-18 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.347 Å) | | Cite: | Organometallic and radical intermediates reveal mechanism of diphthamide biosynthesis.
Science, 359, 2018
|
|
6BXO
 
 | | Crystal structure of Candidatus Methanoperedens nitroreducens Dph2 with 4Fe-4S cluster and SAH | | Descriptor: | Diphthamide biosynthesis enzyme Dph2, IRON/SULFUR CLUSTER, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Fenwick, M.K, Torelli, A.T, Zhang, Y, Dong, M, Kathiresan, V, Carantoa, J.D, Dzikovski, B, Lancaster, K.M, Freed, J.H, Hoffman, B.M, Lin, H, Ealick, S.E. | | Deposit date: | 2017-12-18 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.655 Å) | | Cite: | Organometallic and radical intermediates reveal mechanism of diphthamide biosynthesis.
Science, 359, 2018
|
|
5HFU
 
 | | Crystal Structure of Human Hexokinase 2 with cmpd 27, a 2-amido-6-benzenesulfonamide glucosamine | | Descriptor: | Hexokinase-2, ~{N}-[(2~{S},3~{R},4~{R},5~{S},6~{R})-6-[[(4-cyanophenyl)sulfonylamino]methyl]-2,4,5-tris(oxidanyl)oxan-3-yl]-3-phenyl-benzamide | | Authors: | Campobasso, N, Zhao, B, Smallwood, A. | | Deposit date: | 2016-01-07 | | Release date: | 2016-03-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.923 Å) | | Cite: | Discovery of a Novel 2,6-Disubstituted Glucosamine Series of Potent and Selective Hexokinase 2 Inhibitors.
Acs Med.Chem.Lett., 7, 2016
|
|
5HEX
 
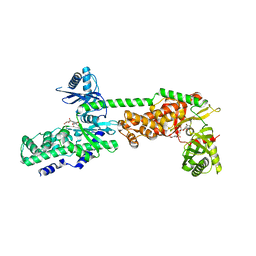 | | Crystal Structure of Human Hexokinase 2 with cmpd 30, a 2-amino-6-benzenesulfonamide glucosamine | | Descriptor: | 2-[(3-bromobenzene-1-carbonyl)amino]-6-{[(4-carboxy-5-methylfuran-2-yl)sulfonyl]amino}-2,6-dideoxy-alpha-D-glucopyranos e, Hexokinase-2 | | Authors: | Campobasso, N, Zhao, B, Smallwood, A. | | Deposit date: | 2016-01-06 | | Release date: | 2016-03-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.734 Å) | | Cite: | Discovery of a Novel 2,6-Disubstituted Glucosamine Series of Potent and Selective Hexokinase 2 Inhibitors.
Acs Med.Chem.Lett., 7, 2016
|
|
5HG1
 
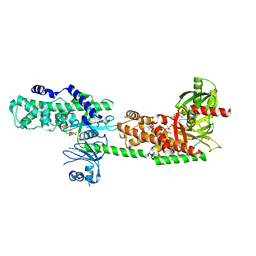 | | Crystal Structure of Human Hexokinase 2 with cmpd 1, a C-2-substituted glucosamine | | Descriptor: | 2-deoxy-2-{[(2E)-3-(3,4-dichlorophenyl)prop-2-enoyl]amino}-alpha-D-glucopyranose, 6-O-phosphono-beta-D-glucopyranose, CITRATE ANION, ... | | Authors: | Campobasso, N, Zhao, B, Smallwood, A. | | Deposit date: | 2016-01-07 | | Release date: | 2016-03-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Discovery of a Novel 2,6-Disubstituted Glucosamine Series of Potent and Selective Hexokinase 2 Inhibitors.
Acs Med.Chem.Lett., 7, 2016
|
|
3KKV
 
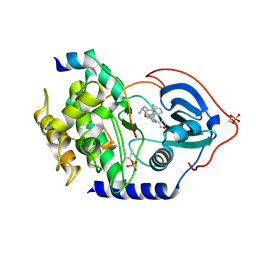 | | Structure of PKA with a protein Kinase B-selective inhibitor. | | Descriptor: | (2S)-1-{[6-furan-3-yl-5-(3-methyl-2H-indazol-5-yl)pyridin-3-yl]oxy}-3-(1H-indol-3-yl)propan-2-amine, PKI kinase inhibitor, cAMP-dependent protein kinase catalytic subunit alpha | | Authors: | Elkins, P.A, Concha, N.O. | | Deposit date: | 2009-11-06 | | Release date: | 2010-12-22 | | Last modified: | 2014-12-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of PKA with a protein Kinase B-selective inhibitor.
To be Published
|
|
6A73
 
 | | Complex structure of CSN2 with IP6 | | Descriptor: | COP9 signalosome complex subunit 2,Endolysin, INOSITOL HEXAKISPHOSPHATE, SULFATE ION | | Authors: | Liu, L, Li, D, Rao, F, Wang, T. | | Deposit date: | 2018-07-02 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.447 Å) | | Cite: | Basis for metabolite-dependent Cullin-RING ligase deneddylation by the COP9 signalosome.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8JMZ
 
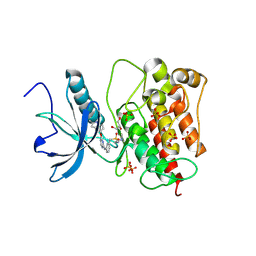 | | FGFR1 kinase domain with sulfatinib | | Descriptor: | Fibroblast growth factor receptor 1, GLYCEROL, N-(2-(dimethylamino)ethyl)-1-(3-((4-((2-methyl-1H-indol-5-yl)oxy)pyrimidin-2-yl)amino)phenyl)methanesulfonamide, ... | | Authors: | Chen, X.J, Lin, Q.M, Chen, Y.H. | | Deposit date: | 2023-06-05 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.988 Å) | | Cite: | Structural basis and selectivity of sulfatinib binding to FGFR and CSF-1R.
Commun Chem, 7, 2024
|
|
8JOT
 
 | | Crystal structure of CSF-1R kinase domain with sulfatinib | | Descriptor: | GLYCEROL, Macrophage colony-stimulating factor 1 receptor, N-(2-(dimethylamino)ethyl)-1-(3-((4-((2-methyl-1H-indol-5-yl)oxy)pyrimidin-2-yl)amino)phenyl)methanesulfonamide | | Authors: | Lin, Q.M, Chen, X.J, Chen, Y.H. | | Deposit date: | 2023-06-08 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural basis and selectivity of sulfatinib binding to FGFR and CSF-1R.
Commun Chem, 7, 2024
|
|
3N9M
 
 | | ceKDM7A from C.elegans, alone | | Descriptor: | FE (II) ION, Putative uncharacterized protein, ZINC ION | | Authors: | Yang, Y, Hu, L, Wang, P, Hou, H, Chen, C.D, Xu, Y. | | Deposit date: | 2010-05-31 | | Release date: | 2010-06-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.493 Å) | | Cite: | Structural insights into a dual-specificity histone demethylase ceKDM7A from Caenorhabditis elegans
Cell Res., 20, 2010
|
|
3N9P
 
 | | ceKDM7A from C.elegans, complex with H3K4me3K27me2 peptide and NOG | | Descriptor: | FE (II) ION, Histone H3 peptide, N-OXALYLGLYCINE, ... | | Authors: | Yang, Y, Hu, L, Wang, P, Hou, H, Chen, C.D, Xu, Y. | | Deposit date: | 2010-05-31 | | Release date: | 2010-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.388 Å) | | Cite: | Structural insights into a dual-specificity histone demethylase ceKDM7A from Caenorhabditis elegans
Cell Res., 20, 2010
|
|
3N9O
 
 | | ceKDM7A from C.elegans, complex with H3K4me3 peptide, H3K9me2 peptide and NOG | | Descriptor: | FE (II) ION, Histone H3 peptide, N-OXALYLGLYCINE, ... | | Authors: | Yang, Y, Hu, L, Wang, P, Hou, H, Chen, C.D, Xu, Y. | | Deposit date: | 2010-05-31 | | Release date: | 2010-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.309 Å) | | Cite: | Structural insights into a dual-specificity histone demethylase ceKDM7A from Caenorhabditis elegans
Cell Res., 20, 2010
|
|
