6IGO
 
 | | Crystal structure of myelin protein zero-like protein 1 (MPZL1) | | Descriptor: | Myelin protein zero-like protein 1 | | Authors: | Yu, T. | | Deposit date: | 2018-09-25 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.746 Å) | | Cite: | Structural and biochemical studies of the extracellular domain of Myelin protein zero-like protein 1
Biochem. Biophys. Res. Commun., 506, 2018
|
|
6IGT
 
 | | MPZL1 mutant - V145G, Q146K, P147T and G148S | | Descriptor: | Myelin protein zero-like protein 1 | | Authors: | Yu, T. | | Deposit date: | 2018-09-26 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.404 Å) | | Cite: | Structural and biochemical studies of the extracellular domain of Myelin protein zero-like protein 1
Biochem. Biophys. Res. Commun., 506, 2018
|
|
6IGW
 
 | | MPZL1 mutant - S86G, V145G, Q146K,P147T,G148S | | Descriptor: | Myelin protein zero-like protein 1 | | Authors: | Yu, T. | | Deposit date: | 2018-09-26 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.979 Å) | | Cite: | Structural and biochemical studies of the extracellular domain of Myelin protein zero-like protein 1
Biochem. Biophys. Res. Commun., 506, 2018
|
|
6IWR
 
 | | Crystal structure of GalNAc-T7 with UDP, GalNAc and Mn2+ | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MANGANESE (II) ION, N-acetylgalactosaminyltransferase 7, ... | | Authors: | Yu, C, Yin, Y.X. | | Deposit date: | 2018-12-06 | | Release date: | 2019-02-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Structural basis of carbohydrate transfer activity of UDP-GalNAc: Polypeptide N-acetylgalactosaminyltransferase 7.
Biochem. Biophys. Res. Commun., 510, 2019
|
|
6IWQ
 
 | | Crystal structure of GalNAc-T7 with Mn2+ | | Descriptor: | MANGANESE (II) ION, N-acetylgalactosaminyltransferase 7 | | Authors: | Yu, C, Yin, Y.X. | | Deposit date: | 2018-12-06 | | Release date: | 2019-02-06 | | Last modified: | 2019-02-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural basis of carbohydrate transfer activity of UDP-GalNAc: Polypeptide N-acetylgalactosaminyltransferase 7.
Biochem. Biophys. Res. Commun., 510, 2019
|
|
6JXT
 
 | |
6JX0
 
 | |
6JWL
 
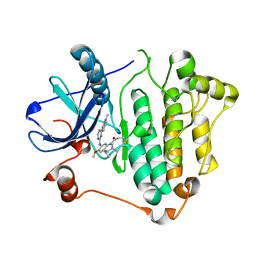 | | Crystal structure of EGFR 696-1022 L858R in complex with AZD9291 | | Descriptor: | Epidermal growth factor receptor, N-(2-{[2-(dimethylamino)ethyl](methyl)amino}-4-methoxy-5-{[4-(1-methyl-1H-indol-3-yl)pyrimidin-2-yl]amino}phenyl)prop-2-enamide | | Authors: | Yun, C.H, Zhu, S.J, Yan, X.E. | | Deposit date: | 2019-04-21 | | Release date: | 2020-04-22 | | Last modified: | 2020-11-04 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | Structural Basis of AZD9291 Selectivity for EGFR T790M.
J.Med.Chem., 63, 2020
|
|
6JX4
 
 | |
5XDK
 
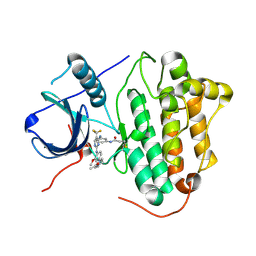 | | Crystal structure of EGFR 696-1022 T790M in complex with CO-1686 | | Descriptor: | Epidermal growth factor receptor, N-[3-[[2-[[4-(4-ethanoylpiperazin-1-yl)-2-methoxy-phenyl]amino]-5-(trifluoromethyl)pyrimidin-4-yl]amino]phenyl]prop-2-enamide | | Authors: | Yan, X.E, Yun, C.H. | | Deposit date: | 2017-03-28 | | Release date: | 2017-12-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.346 Å) | | Cite: | Structural basis of mutant-selectivity and drug-resistance related to CO-1686.
Oncotarget, 8, 2017
|
|
5XDL
 
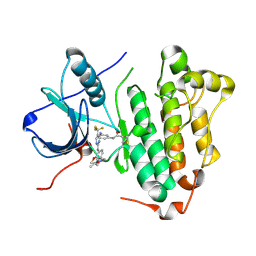 | | Crystal structure of EGFR 696-1022 L858R in complex with CO-1686 | | Descriptor: | Epidermal growth factor receptor, N-[3-[[2-[[4-(4-ethanoylpiperazin-1-yl)-2-methoxy-phenyl]amino]-5-(trifluoromethyl)pyrimidin-4-yl]amino]phenyl]prop-2-enamide | | Authors: | Yan, X.E, Zhu, S.J, Yun, C.H. | | Deposit date: | 2017-03-28 | | Release date: | 2017-12-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of mutant-selectivity and drug-resistance related to CO-1686.
Oncotarget, 8, 2017
|
|
7X1G
 
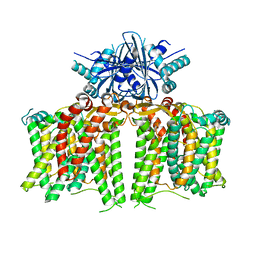 | |
7X1H
 
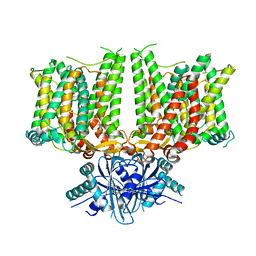 | |
7X1J
 
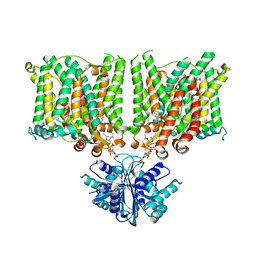 | | Cryo-EM structure of human BTR1 in the outward-facing state in the presence of NH4Cl. | | Descriptor: | Isoform 1 of Solute carrier family 4 member 11, [(2R)-1-octadecanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phospho ryl]oxy-propan-2-yl] (8Z)-icosa-5,8,11,14-tetraenoate | | Authors: | Yin, Y, Lu, Y, Zuo, P. | | Deposit date: | 2022-02-24 | | Release date: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structural insights into the conformational changes of BTR1/SLC4A11 in complex with PIP 2.
Nat Commun, 14, 2023
|
|
7X1I
 
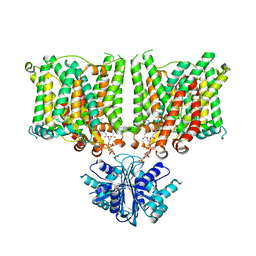 | | Cryo-EM structure of human BTR1 in the outward-facing state. | | Descriptor: | Isoform 1 of Solute carrier family 4 member 11, [(2R)-1-octadecanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phospho ryl]oxy-propan-2-yl] (8Z)-icosa-5,8,11,14-tetraenoate | | Authors: | Yin, Y, Lu, Y, Zuo, P. | | Deposit date: | 2022-02-24 | | Release date: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Structural insights into the conformational changes of BTR1/SLC4A11 in complex with PIP 2.
Nat Commun, 14, 2023
|
|
4MQ2
 
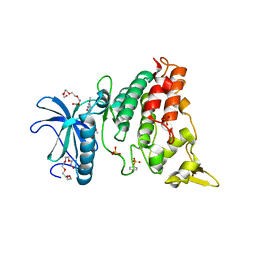 | | The crystal structure of DYRK1a with a bound pyrido[2,3-d]pyrimidine inhibitor | | Descriptor: | Dual specificity tyrosine-phosphorylation-regulated kinase 1A, PENTAETHYLENE GLYCOL, SULFATE ION, ... | | Authors: | Lukacs, C.M, Janson, C.A, Garvie, C, Liang, L. | | Deposit date: | 2013-09-15 | | Release date: | 2013-12-11 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Pyrido[2,3-d]pyrimidines: Discovery and preliminary SAR of a novel series of DYRK1B and DYRK1A inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4MQ1
 
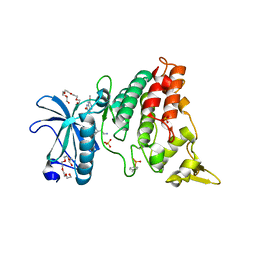 | | The crystal structure of DYRK1a with a bound pyrido[2,3-d]pyrimidine inhibitor | | Descriptor: | Dual specificity tyrosine-phosphorylation-regulated kinase 1A, N-(5-{[(1R)-3-amino-1-(3-chlorophenyl)propyl]carbamoyl}-2-chlorophenyl)-2-methoxy-7-oxo-7,8-dihydropyrido[2,3-d]pyrimidine-6-carboxamide, PENTAETHYLENE GLYCOL, ... | | Authors: | Lukacs, C.M, Janson, C.A, Garvie, C, Liang, L. | | Deposit date: | 2013-09-15 | | Release date: | 2013-12-11 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Pyrido[2,3-d]pyrimidines: Discovery and preliminary SAR of a novel series of DYRK1B and DYRK1A inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
