3B5L
 
 | |
3HOJ
 
 | |
2N3X
 
 | |
2N4H
 
 | |
2N4G
 
 | |
4XFO
 
 | |
4TL5
 
 | | Crystal Structure of Human Transthyretin Ser85Pro Mutant | | Descriptor: | CHLORIDE ION, SODIUM ION, Transthyretin | | Authors: | Saelices, L, Cascio, D, Sawaya, M, Eisenberg, D.S. | | Deposit date: | 2014-05-28 | | Release date: | 2015-10-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Uncovering the Mechanism of Aggregation of Human Transthyretin.
J.Biol.Chem., 290, 2015
|
|
4TKW
 
 | |
4TL4
 
 | |
4TNE
 
 | | Crystal Structure of Human Transthyretin Thr119Tyr Mutant | | Descriptor: | GLYCEROL, Transthyretin | | Authors: | Saelices, L, Cascio, D, Sawaya, M, Eisenberg, D.S. | | Deposit date: | 2014-06-03 | | Release date: | 2015-10-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Uncovering the Mechanism of Aggregation of Human Transthyretin.
J.Biol.Chem., 290, 2015
|
|
4TLS
 
 | | Crystal Structure of Human Transthyretin Glu92Pro Mutant | | Descriptor: | GLYCEROL, PHOSPHATE ION, SODIUM ION, ... | | Authors: | Saelices, L, Cascio, D, Sawaya, M, Eisenberg, D.S. | | Deposit date: | 2014-05-30 | | Release date: | 2015-10-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Uncovering the Mechanism of Aggregation of Human Transthyretin.
J.Biol.Chem., 290, 2015
|
|
4TLT
 
 | | Crystal structure of human transthyretin | | Descriptor: | MAGNESIUM ION, Transthyretin | | Authors: | Saelices, L, Cascio, D, Sawaya, M, Eisenberg, D.S. | | Deposit date: | 2014-05-30 | | Release date: | 2015-10-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Uncovering the Mechanism of Aggregation of Human Transthyretin.
J.Biol.Chem., 290, 2015
|
|
4TLK
 
 | | Crystal Structure of Human Transthyretin Ser85Pro/Glu92Pro Mutant | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, SODIUM ION, ... | | Authors: | Saelices, L, Cascio, D, Sawaya, M, Eisenberg, D.S. | | Deposit date: | 2014-05-30 | | Release date: | 2015-10-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Uncovering the Mechanism of Aggregation of Human Transthyretin.
J.Biol.Chem., 290, 2015
|
|
4TM9
 
 | |
4TJX
 
 | | Crystal structure of protease-associated domain of Arabidopsis VSR1 in complex with aleurain peptide | | Descriptor: | Aleurain peptide, Vacuolar-sorting receptor 1 | | Authors: | Luo, F, Fong, Y.H, Jiang, L.W, Wong, K.B. | | Deposit date: | 2014-05-25 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | How vacuolar sorting receptor proteins interact with their cargo proteins: crystal structures of apo and cargo-bound forms of the protease-associated domain from an Arabidopsis vacuolar sorting receptor.
Plant Cell, 26, 2014
|
|
4TJV
 
 | | Crystal structure of protease-associated domain of Arabidopsis vacuolar sorting receptor 1 | | Descriptor: | IODIDE ION, Vacuolar-sorting receptor 1 | | Authors: | Luo, F, Fong, Y.H, Jiang, L.W, Wong, K.B. | | Deposit date: | 2014-05-25 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | How vacuolar sorting receptor proteins interact with their cargo proteins: crystal structures of apo and cargo-bound forms of the protease-associated domain from an Arabidopsis vacuolar sorting receptor.
Plant Cell, 26, 2014
|
|
4RIL
 
 | | Structure of the amyloid forming segment, GAVVTGVTAVA, from the NAC domain of Parkinson's disease protein alpha-synuclein, residues 68-78, determined by electron diffraction | | Descriptor: | Alpha-synuclein | | Authors: | Rodriguez, J.A, Ivanova, M, Sawaya, M.R, Cascio, D, Reyes, F, Shi, D, Johnson, L, Guenther, E, Sangwan, S, Hattne, J, Nannenga, B, Brewster, A.S, Messerschmidt, M, Boutet, S, Sauter, N.K, Gonen, T, Eisenberg, D.S. | | Deposit date: | 2014-10-06 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-20 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.43 Å) | | Cite: | Structure of the toxic core of alpha-synuclein from invisible crystals.
Nature, 525, 2015
|
|
8TBF
 
 | | Tricomplex of RMC-7977, KRAS WT, and CypA | | Descriptor: | (1R,5S,6r)-N-[(1P,7S,9S,13S,20M)-20-{5-(4-cyclopropylpiperazin-1-yl)-2-[(1S)-1-methoxyethyl]pyridin-3-yl}-21-ethyl-17,17-dimethyl-8,14-dioxo-15-oxa-4-thia-9,21,27,28-tetraazapentacyclo[17.5.2.1~2,5~.1~9,13~.0~22,26~]octacosa-1(24),2,5(28),19,22,25-hexaen-7-yl]-3-oxabicyclo[3.1.0]hexane-6-carboxamide, 1,2-ETHANEDIOL, GTPase KRas, ... | | Authors: | Tomlinson, A.C.A, Chen, A, Knox, J.E, Yano, J.K. | | Deposit date: | 2023-06-28 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Concurrent inhibition of oncogenic and wild-type RAS-GTP for cancer therapy.
Nature, 2024
|
|
7V29
 
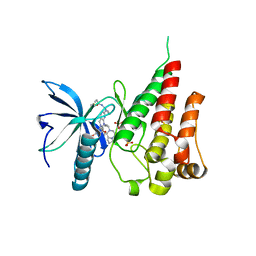 | | Crystal structure of FGFR4 with a dual-warhead covalent inhhibitor | | Descriptor: | Fibroblast growth factor receptor 4, N-[2-[[3-(3,5-dimethoxyphenyl)-2-oxidanylidene-1-[3-(4-propanoylpiperazin-1-yl)propyl]-4H-pyrimido[4,5-d]pyrimidin-7-yl]amino]phenyl]propanamide, SULFATE ION | | Authors: | Chen, X.J, Jiang, L.Y, Dai, S.Y, Qu, L.Z, Chen, Y.H. | | Deposit date: | 2021-08-07 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.983 Å) | | Cite: | Structure-based design of a dual-warhead covalent inhibitor of FGFR4.
Commun Chem, 5, 2022
|
|
6XVD
 
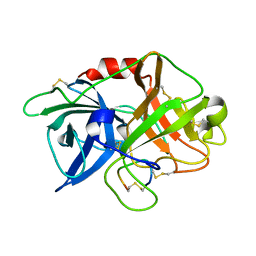 | | Crystal structure of complex of urokinase and a upain-1 variant(W3F) in pH7.4 condition | | Descriptor: | Urokinase-type plasminogen activator, upain-1-W3F | | Authors: | Xue, G.P, Xie, X, Zhou, Y, Yuan, C, Huang, M.D, Jiang, L.G. | | Deposit date: | 2020-01-21 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Insight to the residue in P2 position prevents the peptide inhibitor from being hydrolyzed by serine proteases.
Biosci.Biotechnol.Biochem., 84, 2020
|
|
4RIK
 
 | | Amyloid forming segment, AVVTGVTAV, from the NAC domain of Parkinson's disease protein alpha-synuclein, residues 69-77 | | Descriptor: | Alpha-synuclein | | Authors: | Guenther, E.L, Sawaya, M.R, Ivanova, M, Eisenberg, D.S. | | Deposit date: | 2014-10-06 | | Release date: | 2015-08-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.854 Å) | | Cite: | Structure of the toxic core of alpha-synuclein from invisible crystals.
Nature, 525, 2015
|
|
4RP7
 
 | |
4RP6
 
 | |
8XLO
 
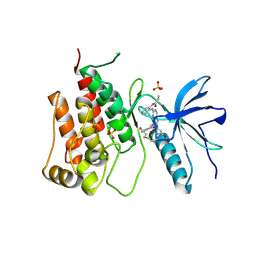 | |
8XLQ
 
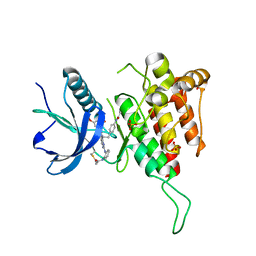 | |
