2NRF
 
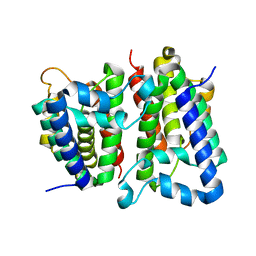 | | Crystal Structure of GlpG, a Rhomboid family intramembrane protease | | Descriptor: | Protein GlpG | | Authors: | Wu, Z, Yan, N, Feng, L, Yan, H, Gu, L, Shi, Y. | | Deposit date: | 2006-11-02 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural analysis of a rhomboid family intramembrane protease reveals a gating mechanism for substrate entry.
Nat.Struct.Mol.Biol., 13, 2006
|
|
5CTG
 
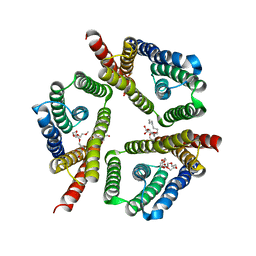 | | The 3.1 A resolution structure of a eukaryotic SWEET transporter | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, Bidirectional sugar transporter SWEET2b, ... | | Authors: | Tao, Y, Perry, K, Feng, L. | | Deposit date: | 2015-07-24 | | Release date: | 2015-10-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.103 Å) | | Cite: | Structure of a eukaryotic SWEET transporter in a homotrimeric complex.
Nature, 527, 2015
|
|
3WE5
 
 | |
6C5R
 
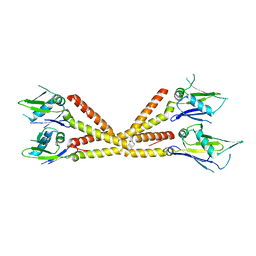 | | Crystal structure of the soluble domain of the mitochondrial calcium uniporter | | Descriptor: | calcium uniporter | | Authors: | Fan, C, Fan, M, Fastman, N, Zhang, J, Feng, L. | | Deposit date: | 2018-01-16 | | Release date: | 2018-07-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.09608316 Å) | | Cite: | X-ray and cryo-EM structures of the mitochondrial calcium uniporter.
Nature, 559, 2018
|
|
6C5W
 
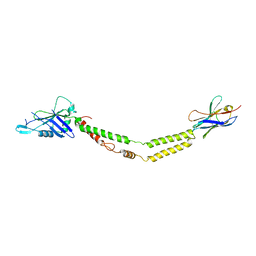 | | Crystal structure of the mitochondrial calcium uniporter | | Descriptor: | CALCIUM ION, calcium uniporter, nanobody | | Authors: | Fan, C, Fan, M, Fastman, N, Zhang, J, Feng, L. | | Deposit date: | 2018-01-17 | | Release date: | 2018-07-11 | | Last modified: | 2019-04-24 | | Method: | X-RAY DIFFRACTION (3.10010242 Å) | | Cite: | X-ray and cryo-EM structures of the mitochondrial calcium uniporter.
Nature, 559, 2018
|
|
5UHS
 
 | | Structure of a SemiSWEET D57A mutant | | Descriptor: | MONOVACCENIN, Sugar transporter SemiSWEET, beta-D-glucopyranose | | Authors: | Fastman, N.M, Feng, L. | | Deposit date: | 2017-01-11 | | Release date: | 2017-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanism of Substrate Translocation in an Alternating Access Transporter.
Cell, 169, 2017
|
|
5UHQ
 
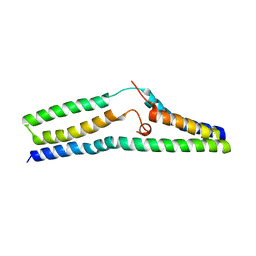 | | Structure of a SemiSWEET Q20A mutant | | Descriptor: | Sugar transporter SemiSWEET | | Authors: | Fastman, N.M, Feng, L. | | Deposit date: | 2017-01-11 | | Release date: | 2017-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Mechanism of Substrate Translocation in an Alternating Access Transporter.
Cell, 169, 2017
|
|
7YLQ
 
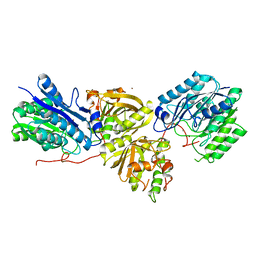 | | Crystal structure of Microcystinase C from Sphingomonas sp. ACM-3962 at 2.6 A resolution | | Descriptor: | CALCIUM ION, Microcystinase C, ZINC ION | | Authors: | Guo, X, Li, Z, Ding, W, Yin, P, Feng, L. | | Deposit date: | 2022-07-26 | | Release date: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Crystal structure of Microcystin C from Sphingomonas sp. ACM-3962 at 2.6 A resolution
To Be Published
|
|
7DYN
 
 | | Phosphorylation of MHC I peptide | | Descriptor: | ARG-ARG-PHE-SEP-ARG-SEP-PRO-ILE-ARG-ARG, Beta-2-microglobulin, MHC class I antigen | | Authors: | Sun, M.W, Feng, L, Qi, J.X, Liu, W.J. | | Deposit date: | 2021-01-22 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phosphosite-dependent presentation of dual phosphorylated peptides by MHC class I molecules.
Iscience, 25, 2022
|
|
7CIS
 
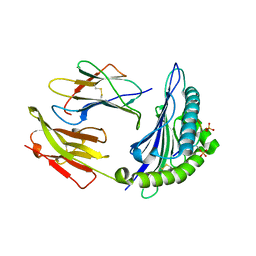 | | Peptide modification of MHC class I molecules | | Descriptor: | ARG-ARG-PHE-SEP-ARG-SEP-PRO-ILE-ARG, Beta-2-microglobulin, MHC class I antigen | | Authors: | Sun, M.W, Feng, L, Qi, J.X, Liu, W.J, Gao, G.F. | | Deposit date: | 2020-07-08 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Phosphosite-dependent presentation of dual phosphorylated peptides by MHC class I molecules.
Iscience, 25, 2022
|
|
7CIR
 
 | | Peptide phosphorylation modification of MHC class I molecules | | Descriptor: | ARG-ARG-PHE-SEP-ARG-SER-PRO-ILE-ARG-ARG, Beta-2-microglobulin, MHC class I antigen | | Authors: | Sun, M.W, Feng, L, Qi, J.X, Liu, W.J, Gao, G.F. | | Deposit date: | 2020-07-08 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Phosphosite-dependent presentation of dual phosphorylated peptides by MHC class I molecules.
Iscience, 25, 2022
|
|
7CIQ
 
 | | Phosphorylation modification of MHC I polypeptide | | Descriptor: | ARG-ARG-PHE-SER-ARG-SER-PRO-ILE-ARG-ARG, Beta-2-microglobulin, MHC class I antigen | | Authors: | Sun, M.W, Feng, L, Qi, J.X, Liu, W.J, Gao, G.F. | | Deposit date: | 2020-07-08 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Phosphosite-dependent presentation of dual phosphorylated peptides by MHC class I molecules.
Iscience, 25, 2022
|
|
3PUP
 
 | | Structure of Glycogen Synthase Kinase 3 beta (GSK3B) in complex with a ruthenium octasporine ligand (OS1) | | Descriptor: | Glycogen synthase kinase-3 beta, Ruthenium octasporine | | Authors: | Filippakopoulos, P, Kraling, K, Essen, L.O, Meggers, E, Knapp, S. | | Deposit date: | 2010-12-06 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structurally sophisticated octahedral metal complexes as highly selective protein kinase inhibitors.
J.Am.Chem.Soc., 133, 2011
|
|
5JL4
 
 | |
8IN7
 
 | | Crystal structure of UGT74AN3-UDP | | Descriptor: | Glycosyltransferase, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Feng, L, Wei, H. | | Deposit date: | 2023-03-08 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of UGT74AN3-UDP
To Be Published
|
|
4RZE
 
 | | Crystal Structure Analysis of the NUR77 Ligand Binding Domain, L437W,D594E mutant | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Fengwei, L, Xuyang, T, Anzhong, L, Li, L, Yuan, L, Hangzi, C, Qiao, W, Tianwei, L. | | Deposit date: | 2014-12-21 | | Release date: | 2015-03-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Impeding the interaction between Nur77 and p38 reduces LPS-induced inflammation.
Nat.Chem.Biol., 11, 2015
|
|
5D06
 
 | |
5H19
 
 | | EED in complex with PRC2 allosteric inhibitor EED162 | | Descriptor: | 5-(furan-2-ylmethylamino)-9-(phenylmethyl)-8,10-dihydro-7H-[1,2,4]triazolo[3,4-a][2,7]naphthyridine-6-carbonitrile, Histone-lysine N-methyltransferase EZH2, Polycomb protein EED | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H. | | Deposit date: | 2016-10-08 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and Molecular Basis of a Diverse Set of Polycomb Repressive Complex 2 Inhibitors Recognition by EED
PLoS ONE, 12, 2017
|
|
5KRS
 
 | | HIV-1 Integrase Catalytic Core Domain in Complex with an Allosteric Inhibitor, 3-(1H-pyrrol-1-yl)-2-thiophenecarboxylic acid | | Descriptor: | 3-pyrrol-1-ylthiophene-2-carboxylic acid, DIMETHYL SULFOXIDE, Integrase | | Authors: | Patel, D, Bauman, J.D, Arnold, E. | | Deposit date: | 2016-07-07 | | Release date: | 2016-09-28 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A New Class of Allosteric HIV-1 Integrase Inhibitors Identified by Crystallographic Fragment Screening of the Catalytic Core Domain.
J.Biol.Chem., 291, 2016
|
|
4WYU
 
 | |
3A9U
 
 | |
4WYT
 
 | |
3A9V
 
 | |
7QJU
 
 | | EED in complex with PRC2 allosteric inhibitor compound 7 | | Descriptor: | CHLORIDE ION, Histone-lysine N-methyltransferase EZH2, N-(2,3-dihydro-1-benzofuran-4-ylmethyl)-8-[4-[(dimethylamino)methyl]phenyl]-[1,2,4]triazolo[4,3-c]pyrimidin-5-amine, ... | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H, Scheufler, C. | | Deposit date: | 2021-12-17 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of the Clinical Candidate MAK683: An EED-Directed, Allosteric, and Selective PRC2 Inhibitor for the Treatment of Advanced Malignancies.
J.Med.Chem., 65, 2022
|
|
7QJG
 
 | | EED in complex with PRC2 allosteric inhibitor compound 6 | | Descriptor: | CHLORIDE ION, Histone-lysine N-methyltransferase EZH2, N-(2,3-dihydro-1-benzofuran-7-ylmethyl)-8-[4-[(dimethylamino)methyl]phenyl]-[1,2,4]triazolo[4,3-c]pyrimidin-5-amine, ... | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H, Scheufler, C. | | Deposit date: | 2021-12-16 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of the Clinical Candidate MAK683: An EED-Directed, Allosteric, and Selective PRC2 Inhibitor for the Treatment of Advanced Malignancies.
J.Med.Chem., 65, 2022
|
|
