2L8R
 
 | |
2L4N
 
 | |
2LQ7
 
 | |
2LGR
 
 | |
2LC6
 
 | |
2LVX
 
 | |
2MBF
 
 | |
2MG4
 
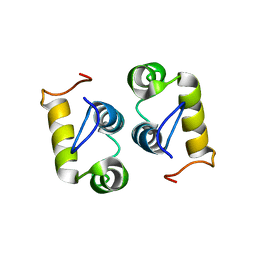 | | Computational design and experimental verification of a symmetric protein homodimer | | Descriptor: | Computational designed homodimer | | Authors: | Mou, Y, Huang, P.S, Hsu, F.C, Huang, S.J, Mayo, S.L. | | Deposit date: | 2013-10-26 | | Release date: | 2015-04-08 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Computational design and experimental verification of a symmetric protein homodimer.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2LWX
 
 | |
2MP1
 
 | |
2N1H
 
 | |
2N55
 
 | |
2OJ7
 
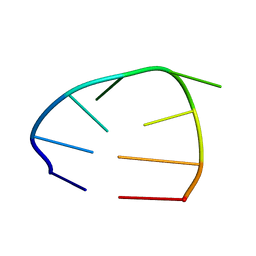 | | NMR structure of the UGUU tetraloop of Duck Epsilon apical stem loop | | Descriptor: | 5'-R(P*GP*CP*UP*GP*UP*UP*GP*U)-3' | | Authors: | Girard, F.C, Ottink, O.M, Ampt, K.A.M, Tessari, M, Wijmenga, S.S. | | Deposit date: | 2007-01-12 | | Release date: | 2007-05-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Thermodynamics and NMR studies on Duck, Heron and Human HBV encapsidation signals.
Nucleic Acids Res., 35, 2007
|
|
2QED
 
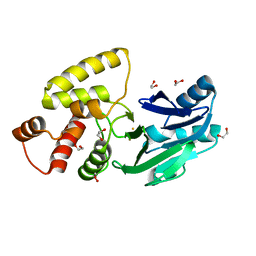 | | Crystal structure of Salmonella thyphimurium LT2 glyoxalase II | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, Hydroxyacylglutathione hydrolase | | Authors: | Leite, N.R, Campos Bermudez, V.A, Krogh, R, Oliva, G, Soncini, F.C, Vila, A.J. | | Deposit date: | 2007-06-25 | | Release date: | 2007-10-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Biochemical and Structural Characterization of Salmonella typhimurium Glyoxalase II: New Insights into Metal Ion Selectivity
Biochemistry, 46, 2007
|
|
2ROD
 
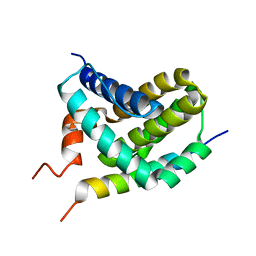 | | Solution Structure of MCL-1 Complexed with NoxaA | | Descriptor: | Induced myeloid leukemia cell differentiation protein Mcl-1 homolog, Noxa | | Authors: | Day, C.L, Smits, C, Fan, F.C, Lee, E.F, Fairlie, W.D, Hinds, M.G. | | Deposit date: | 2008-03-17 | | Release date: | 2008-07-08 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structure of the BH3 Domains from the p53-Inducible BH3-Only Proteins Noxa and Puma in Complex with Mcl-1
J.Mol.Biol., 380, 2008
|
|
2ROC
 
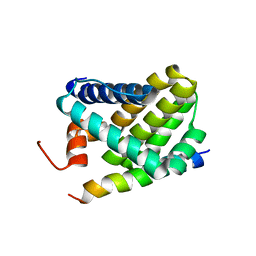 | | Solution structure of Mcl-1 Complexed with Puma | | Descriptor: | Bcl-2-binding component 3, Induced myeloid leukemia cell differentiation protein Mcl-1 homolog | | Authors: | Day, C.L, Smits, C, Fan, F.C, Lee, E.F, Fairlie, W.D, Hinds, M.G. | | Deposit date: | 2008-03-17 | | Release date: | 2008-07-08 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Structure of the BH3 Domains from the p53-Inducible BH3-Only Proteins Noxa and Puma in Complex with Mcl-1
J.Mol.Biol., 380, 2008
|
|
2REM
 
 | |
2RLJ
 
 | | NMR Structure of Ebola fusion peptide in SDS micelles at pH 7 | | Descriptor: | Envelope glycoprotein | | Authors: | Freitas, M.S, Gaspar, L.P, Lorenzoni, M, Almeida, F.C, Tinoco, L.W, Almeida, M.S, Maia, L.F, Degreve, L, Valente, A.P, Silva, J.L. | | Deposit date: | 2007-07-05 | | Release date: | 2007-08-07 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structure of the Ebola fusion peptide in a membrane-mimetic environment and the interaction with lipid rafts.
J.Biol.Chem., 282, 2007
|
|
4F7Z
 
 | | Conformational dynamics of exchange protein directly activated by cAMP | | Descriptor: | GLYCEROL, Rap guanine nucleotide exchange factor 4 | | Authors: | White, M.A, Tsalkova, T.N, Mei, F.C, Liu, T, Woods, V.L, Cheng, X. | | Deposit date: | 2012-05-16 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural analyses of a constitutively active mutant of exchange protein directly activated by cAMP.
Plos One, 7, 2012
|
|
4HSO
 
 | | Crystal structure of S213G variant DAH7PS from Neisseria meningitidis | | Descriptor: | 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase, MANGANESE (II) ION, PHOSPHOENOLPYRUVATE, ... | | Authors: | Cross, P.J, Pietersma, A.L, Allison, T.M, Wilson-Coutts, S.M, Cochrane, F.C, Parker, E.J. | | Deposit date: | 2012-10-30 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Neisseria meningitidis expresses a single 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase that is inhibited primarily by phenylalanine.
Protein Sci., 22, 2013
|
|
4HSN
 
 | | Crystal structure of DAH7PS from Neisseria meningitidis | | Descriptor: | 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase, MANGANESE (II) ION, PHOSPHOENOLPYRUVATE, ... | | Authors: | Cross, P.J, Pietersma, A.L, Allison, T.M, Wilson-Coutts, S.M, Cochrane, F.C, Parker, E.J. | | Deposit date: | 2012-10-30 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Neisseria meningitidis expresses a single 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase that is inhibited primarily by phenylalanine.
Protein Sci., 22, 2013
|
|
4IXX
 
 | | Crystal structure of S213G variant DAH7PS without Tyr bound from Neisseria meningitidis | | Descriptor: | 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase, MANGANESE (II) ION, SULFATE ION | | Authors: | Cross, P.J, Pietersma, A.L, Allison, T.M, Wilson-Coutts, S.M, Cochrane, F.C, Parker, E.J. | | Deposit date: | 2013-01-28 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Neisseria meningitidis expresses a single 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase that is inhibited primarily by phenylalanine.
Protein Sci., 22, 2013
|
|
4JTJ
 
 | | Crystal structure of R117K mutant of 3-deoxy-D-manno-octulosonate 8-phosphate synthase (KDO8PS) from Neisseria meningitidis | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase, CHLORIDE ION, SODIUM ION | | Authors: | Allison, T.M, Cochrane, F.C, Jameson, G.B, Parker, E.J. | | Deposit date: | 2013-03-23 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Examining the Role of Intersubunit Contacts in Catalysis by 3-Deoxy-d-manno-octulosonate 8-Phosphate Synthase.
Biochemistry, 52, 2013
|
|
4JTL
 
 | | Crystal structure of F139G mutant of 3-deoxy-D-manno-octulosonate 8-phosphate synthase (KDO8PS) from Neisseria meningitidis | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase, CHLORIDE ION | | Authors: | Allison, T.M, Cochrane, F.C, Jameson, G.B, Parker, E.J. | | Deposit date: | 2013-03-23 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Examining the Role of Intersubunit Contacts in Catalysis by 3-Deoxy-d-manno-octulosonate 8-Phosphate Synthase.
Biochemistry, 52, 2013
|
|
4JTI
 
 | | Crystal structure of F114R/R117Q/F139G mutant of 3-deoxy-D-manno-octulosonate 8-phosphate synthase (KDO8PS) from Neisseria meningitidis | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase, CHLORIDE ION | | Authors: | Allison, T.M, Cochrane, F.C, Jameson, G.B, Parker, E.J. | | Deposit date: | 2013-03-23 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Examining the Role of Intersubunit Contacts in Catalysis by 3-Deoxy-d-manno-octulosonate 8-Phosphate Synthase.
Biochemistry, 52, 2013
|
|
