7D4G
 
 | |
7DX4
 
 | | The structure of FC08 Fab-hA.CE2-RBD complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Heavy chain of FC08 Fab, ... | | Authors: | Cao, L, Wang, X. | | Deposit date: | 2021-01-18 | | Release date: | 2021-04-21 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A proof of concept for neutralizing antibody-guided vaccine design against SARS-CoV-2.
Natl Sci Rev, 8, 2021
|
|
7EHJ
 
 | | human MTHFD2 in complex with compound 21, cofactor and phosphate. | | Descriptor: | (2S)-2-[[4-[(4-azanyl-6-oxidanyl-pyrimidin-5-yl)carbamoylamino]phenyl]carbonylamino]pentanedioic acid, Bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, mitochondrial, ... | | Authors: | Lee, L.C, Peng, Y.H, Wu, S.Y. | | Deposit date: | 2021-03-29 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Xanthine Derivatives Reveal an Allosteric Binding Site in Methylenetetrahydrofolate Dehydrogenase 2 (MTHFD2).
J.Med.Chem., 64, 2021
|
|
7EHN
 
 | | Human MTHFD2 in complex with compound 21 and 9 | | Descriptor: | (2S)-2-[[4-[(4-azanyl-6-oxidanyl-pyrimidin-5-yl)carbamoylamino]phenyl]carbonylamino]pentanedioic acid, 3-[4-[[1-[(4-chloranyl-1H-indol-2-yl)methyl]-3,7-dimethyl-2,6-bis(oxidanylidene)purin-8-yl]amino]-6-methyl-pyrimidin-2-yl]propanoic acid, Bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, ... | | Authors: | Lee, L.C, Peng, Y.H, Wu, S.Y. | | Deposit date: | 2021-03-30 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Xanthine Derivatives Reveal an Allosteric Binding Site in Methylenetetrahydrofolate Dehydrogenase 2 (MTHFD2).
J.Med.Chem., 64, 2021
|
|
7EHM
 
 | | Human MTHFD2 in complex with compound 21 and 15 | | Descriptor: | (2S)-2-[[4-[(4-azanyl-6-oxidanyl-pyrimidin-5-yl)carbamoylamino]phenyl]carbonylamino]pentanedioic acid, (2S)-2-[[4-[[1-[(3,4-dichlorophenyl)methyl]-3,7-dimethyl-2,6-bis(oxidanylidene)purin-8-yl]amino]phenyl]carbonylamino]pentanedioic acid, Bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, ... | | Authors: | Lee, L.C, Peng, Y.H, Wu, S.Y. | | Deposit date: | 2021-03-30 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Xanthine Derivatives Reveal an Allosteric Binding Site in Methylenetetrahydrofolate Dehydrogenase 2 (MTHFD2).
J.Med.Chem., 64, 2021
|
|
7EHV
 
 | | Human MTHFD2 in complex with compound 21 and 3 | | Descriptor: | (2S)-2-[[4-[(4-azanyl-6-oxidanyl-pyrimidin-5-yl)carbamoylamino]phenyl]carbonylamino]pentanedioic acid, 1-(3,4-dichlorobenzyl)-8-(((1R,4R)-4-hydroxycyclohexyl)amino)-3,7-dimethyl-3,7-dihydro-1H-purine-2,6-dione, Bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, ... | | Authors: | Lee, L.C, Peng, Y.H, Wu, S.Y. | | Deposit date: | 2021-03-30 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Xanthine Derivatives Reveal an Allosteric Binding Site in Methylenetetrahydrofolate Dehydrogenase 2 (MTHFD2).
J.Med.Chem., 64, 2021
|
|
1JKZ
 
 | | NMR Solution Structure of Pisum sativum defensin 1 (Psd1) | | Descriptor: | DEFENSE-RELATED PEPTIDE 1 | | Authors: | Almeida, M.S, Cabral, K.M.S, Kurtenbach, E, Almeida, F.C.L, Valente, A.P. | | Deposit date: | 2001-07-13 | | Release date: | 2002-02-06 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Pisum sativum defensin 1 by high resolution NMR: plant defensins, identical backbone with different mechanisms of action.
J.Mol.Biol., 315, 2002
|
|
1JMO
 
 | | Crystal Structure of the Heparin Cofactor II-S195A Thrombin Complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Baglin, T.P, Carrell, R.W, Esmon, C.T, Huntington, J.A. | | Deposit date: | 2001-07-19 | | Release date: | 2002-08-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of native and thrombin-complexed heparin cofactor II reveal a multistep allosteric mechanism.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1JWW
 
 | | NMR characterization of the N-terminal domain of a potential copper-translocating P-type ATPase from Bacillus subtilis | | Descriptor: | Potential copper-transporting ATPase | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, D'Onofrio, M, Gonnelli, L, Marhuenda-Egea, F, Ruiz-Duenas, F.J. | | Deposit date: | 2001-09-05 | | Release date: | 2002-04-10 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal domain of a potential copper-translocating P-type ATPase from Bacillus subtilis in the apo and Cu(I) loaded states.
J.Mol.Biol., 317, 2002
|
|
6XN0
 
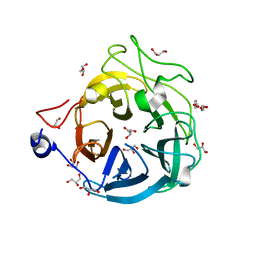 | | Crystal structure of GH43_1 enzyme from Xanthomonas citri | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Morais, M.A.B, Tonoli, C.C.C, Santos, C.R, Murakami, M.T. | | Deposit date: | 2020-07-02 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.709 Å) | | Cite: | Two distinct catalytic pathways for GH43 xylanolytic enzymes unveiled by X-ray and QM/MM simulations.
Nat Commun, 12, 2021
|
|
6XN2
 
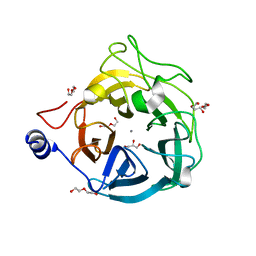 | | Crystal structure of the GH43_1 enzyme from Xanthomonas citri complexed with xylotriose | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Morais, M.A.B, Tonoli, C.C.C, Santos, C.R, Murakami, M.T. | | Deposit date: | 2020-07-02 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.652 Å) | | Cite: | Two distinct catalytic pathways for GH43 xylanolytic enzymes unveiled by X-ray and QM/MM simulations.
Nat Commun, 12, 2021
|
|
6XN1
 
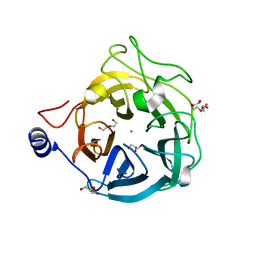 | | Crystal structure of the GH43_1 enzyme from Xanthomonas citri complexed with xylose | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Morais, M.A.B, Tonoli, C.C.C, Santos, C.R, Murakami, M.T. | | Deposit date: | 2020-07-02 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Two distinct catalytic pathways for GH43 xylanolytic enzymes unveiled by X-ray and QM/MM simulations.
Nat Commun, 12, 2021
|
|
5TBO
 
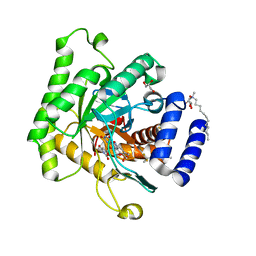 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM421 | | Descriptor: | 2-(1,1-difluoroethyl)-5-methyl-N-[6-(trifluoromethyl)pyridin-3-yl][1,2,4]triazolo[1,5-a]pyrimidin-7-amine, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Deng, X, Phillips, M. | | Deposit date: | 2016-09-12 | | Release date: | 2016-09-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | A Triazolopyrimidine-Based Dihydroorotate Dehydrogenase Inhibitor with Improved Drug-like Properties for Treatment and Prevention of Malaria.
ACS Infect Dis, 2, 2016
|
|
5T3M
 
 | |
5UBM
 
 | | Crystal structure of human C1s in complex with inhibitor gigastasin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C1s subcomponent, Gigastasin | | Authors: | Pang, S.S, Whisstock, J.C. | | Deposit date: | 2016-12-20 | | Release date: | 2017-11-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Structural Basis for Complement Inhibition by Gigastasin, a Protease Inhibitor from the Giant Amazon Leech.
J. Immunol., 199, 2017
|
|
6ZTR
 
 | | Crystal Structure of the anti-human P-Cadherin Fab CQY684 in complex with human P-Cadherin(108-324) | | Descriptor: | CALCIUM ION, CQY684 Fab heavy-chain, CQY684 Fab light-chain, ... | | Authors: | Rondeau, J.M, Lehmann, S. | | Deposit date: | 2020-07-20 | | Release date: | 2021-05-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | PCA062, a P-cadherin Targeting Antibody-Drug Conjugate, Displays Potent Antitumor Activity Against P-cadherin-expressing Malignancies.
Mol.Cancer Ther., 20, 2021
|
|
6ZTB
 
 | | Crystal Structure of human P-Cadherin EC1_EC2 | | Descriptor: | CALCIUM ION, Cadherin-3, SODIUM ION | | Authors: | Rondeau, J.M, Lehmann, S. | | Deposit date: | 2020-07-17 | | Release date: | 2021-05-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | PCA062, a P-cadherin Targeting Antibody-Drug Conjugate, Displays Potent Antitumor Activity Against P-cadherin-expressing Malignancies.
Mol.Cancer Ther., 20, 2021
|
|
6ZTF
 
 | | Crystal Structure of the anti-human P-Cadherin Fab CQY684 | | Descriptor: | CQY684 Fab heavy-chain, CQY684 Fab light-chain | | Authors: | Rondeau, J.M, Lehmann, S. | | Deposit date: | 2020-07-20 | | Release date: | 2021-05-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | PCA062, a P-cadherin Targeting Antibody-Drug Conjugate, Displays Potent Antitumor Activity Against P-cadherin-expressing Malignancies.
Mol.Cancer Ther., 20, 2021
|
|
5VOK
 
 | | Crystal structure of the C7orf59-HBXIP dimer | | Descriptor: | Ragulator complex protein LAMTOR4, Ragulator complex protein LAMTOR5 | | Authors: | Rasheed, N, Nascimento, A.F.Z, Bar-Peled, L, Shen, K, Sabatini, D.M, Aparicio, R, Smetana, J.H.C. | | Deposit date: | 2017-05-03 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | C7orf59/Lamtor4 phosphorylation and structural flexibility modulate Ragulator assembly.
Febs Open Bio, 2019
|
|
1Q4R
 
 | | Gene Product of At3g17210 from Arabidopsis Thaliana | | Descriptor: | MAGNESIUM ION, protein At3g17210 | | Authors: | Phillips Jr, G.N, Bingman, C.A, Johnson, K.A, Smith, D.W, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2003-08-04 | | Release date: | 2003-11-25 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the protein from gene At3g17210 of Arabidopsis thaliana
Proteins, 57, 2004
|
|
2ESE
 
 | | Structure of the SAM domain of Vts1p in complex with RNA | | Descriptor: | 5'-R(*GP*GP*AP*GP*AP*GP*GP*CP*UP*CP*UP*GP*GP*CP*AP*GP*CP*UP*UP*UP*UP*CP*C)-3', Vts1p | | Authors: | Allain, F.H.T. | | Deposit date: | 2005-10-26 | | Release date: | 2006-01-24 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Shape-specific recognition in the structure of the Vts1p SAM domain with RNA.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2EVZ
 
 | |
2ES5
 
 | | Structure of the SRE RNA | | Descriptor: | 5'-R(*GP*GP*AP*GP*AP*GP*GP*CP*UP*CP*UP*GP*GP*CP*AP*GP*CP*UP*UP*UP*UP*CP*C)-3' | | Authors: | Allain, F.H.T. | | Deposit date: | 2005-10-25 | | Release date: | 2006-01-24 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Shape-specific recognition in the structure of the Vts1p SAM domain with RNA.
Nat.Struct.Mol.Biol., 13, 2006
|
|
3G1Z
 
 | | Structure of IDP01693/yjeA, a potential t-RNA synthetase from Salmonella typhimurium | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, PHOSPHATE ION, ... | | Authors: | Singer, A.U, Evdokimova, E, Kudritska, M, Cuff, M.E, Edwards, A.M, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-01-30 | | Release date: | 2009-03-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | PoxA, yjeK, and elongation factor P coordinately modulate virulence and drug resistance in Salmonella enterica.
Mol.Cell, 39, 2010
|
|
3FYO
 
 | | Crystal structure of the triple mutant (N23C/D247E/P249A) of 3-deoxy-D-manno-octulosonate 8-phosphate synthase (KDO8PS) from Neisseria meningitidis | | Descriptor: | 3-deoxy-D-manno-octulosonic acid 8-phosphate synthetase, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Jameson, G.B, Parker, E.J, Cochrane, F.P, Patchett, M.L. | | Deposit date: | 2009-01-22 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Reversing evolution: re-establishing obligate metal ion dependence in a metal-independent KDO8P synthase
J.Mol.Biol., 390, 2009
|
|
