6CY2
 
 | | RNA octamer containing 2'-OMe, 4'Calpha-OMe U. | | Descriptor: | RNA (5'-R(*(CBV)P*GP*AP*AP*(UOA)P*UP*CP*G)-3') | | Authors: | Harp, J.M, Egli, M. | | Deposit date: | 2018-04-04 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for the synergy of 4'- and 2'-modifications on siRNA nuclease resistance, thermal stability and RNAi activity.
Nucleic Acids Res., 46, 2018
|
|
5VR4
 
 | | RNA octamer containing 2'-F-4'-OMe U. | | Descriptor: | COBALT TETRAAMMINE ION, RNA (5'-R(*CP*GP*AP*AP*(UMO)P*UP*CP*G)-3') | | Authors: | Harp, J.M, Egli, M. | | Deposit date: | 2017-05-10 | | Release date: | 2017-10-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 4'-C-Methoxy-2'-deoxy-2'-fluoro Modified Ribonucleotides Improve Metabolic Stability and Elicit Efficient RNAi-Mediated Gene Silencing.
J. Am. Chem. Soc., 139, 2017
|
|
3OZ5
 
 | | S-Methyl Carbocyclic LNA | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*AP*(UMX)P*AP*CP*GP*C)-3'), SPERMINE | | Authors: | Seth, P.R, Allerson, C.A, Berdeja, A, Siwkowski, A, Pallan, P.S, Gaus, H, Prakash, T.P, Watt, A.T, Egli, M, Swayze, E.E. | | Deposit date: | 2010-09-24 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | An exocyclic methylene group acts as a bioisostere of the 2'-oxygen atom in LNA.
J.Am.Chem.Soc., 132, 2010
|
|
3OZ3
 
 | | Vinyl Carbocyclic LNA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*GP*CP*GP*TP*AP*(UVX)P*AP*CP*GP*C)-3') | | Authors: | Seth, P.R, Allerson, C.A, Berdeja, A, Siwkowski, A, Pallan, P.S, Gaus, H, Prakash, T.P, Watt, A.T, Egli, M, Swayze, E.E. | | Deposit date: | 2010-09-24 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | An exocyclic methylene group acts as a bioisostere of the 2'-oxygen atom in LNA.
J.Am.Chem.Soc., 132, 2010
|
|
3OZ4
 
 | | R-Methyl Carbocyclic LNA | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*AP*(URX)P*AP*CP*GP*C)-3') | | Authors: | Seth, P.R, Allerson, C.A, Berdeja, A, Siwkowski, A, Pallan, P.S, Gaus, H, Prakash, T.P, Watt, A.T, Egli, M, Swayze, E.E. | | Deposit date: | 2010-09-24 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | An exocyclic methylene group acts as a bioisostere of the 2'-oxygen atom in LNA.
J.Am.Chem.Soc., 132, 2010
|
|
7LO9
 
 | | RNA dodecamer containing a GNA A residue | | Descriptor: | Chains: A,B,C,D | | Authors: | Harp, J.M, Wawrzak, Z, Egli, M. | | Deposit date: | 2021-02-09 | | Release date: | 2021-12-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Overcoming GNA/RNA base-pairing limitations using isonucleotides improves the pharmacodynamic activity of ESC+ GalNAc-siRNAs.
Nucleic Acids Res., 49, 2021
|
|
2M11
 
 | | Structure of perimidinone-derived synthetic nucleoside paired with guanine in DNA duplex | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*(D3N)P*GP*CP*G)-3') | | Authors: | Kowal, E.A, Lad, R, Pallan, P.S, Muffly, E, Wawrzak, Z, Egli, M, Sturla, S.J, Stone, M.P. | | Deposit date: | 2012-11-09 | | Release date: | 2013-06-12 | | Last modified: | 2013-09-11 | | Method: | SOLUTION NMR | | Cite: | Recognition of O6-benzyl-2'-deoxyguanosine by a perimidinone-derived synthetic nucleoside: a DNA interstrand stacking interaction.
Nucleic Acids Res., 41, 2013
|
|
3S1A
 
 | | Crystal structure of the phosphorylation-site double mutant S431E/T432E of the KaiC circadian clock protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein kinase kaiC, MAGNESIUM ION | | Authors: | Pattanayek, R, Williams, D.W, Rossi, G, Weigand, S, Mori, T, Johnson, C.H, Stewart, P.L, Egli, M. | | Deposit date: | 2011-05-14 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Combined SAXS/EM Based Models of the S. elongatus Post-Translational Circadian Oscillator and its Interactions with the Output His-Kinase SasA.
Plos One, 6, 2011
|
|
5DER
 
 | | RNA oligonucleotide containing (R)-C5'-ME-2'F U | | Descriptor: | RNA oligonucleotide containing (R)-C5'-Me-2'-FU | | Authors: | Harp, J.M, Egli, M. | | Deposit date: | 2015-08-25 | | Release date: | 2016-06-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Duplex Thermodynamic Stability and Enhanced Nuclease Resistance of 5'-C-Methyl Pyrimidine-Modified Oligonucleotides.
J.Org.Chem., 81, 2016
|
|
5D8T
 
 | | RNA octamer containing (S)-5' methyl, 2'-F U. | | Descriptor: | COBALT HEXAMMINE(III), RNA oligonucleotide containing (S)-C5'-Me-2'-FU | | Authors: | Harp, J.M, Egli, M. | | Deposit date: | 2015-08-17 | | Release date: | 2016-06-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural Basis of Duplex Thermodynamic Stability and Enhanced Nuclease Resistance of 5'-C-Methyl Pyrimidine-Modified Oligonucleotides.
J.Org.Chem., 81, 2016
|
|
5DEK
 
 | | RNA octamer containing dT | | Descriptor: | COBALT HEXAMMINE(III), RNA oligonucleotide containing dT | | Authors: | Harp, J.M, Egli, M. | | Deposit date: | 2015-08-25 | | Release date: | 2016-07-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.993 Å) | | Cite: | Structural Basis of Duplex Thermodynamic Stability and Enhanced Nuclease Resistance of 5'-C-Methyl Pyrimidine-Modified Oligonucleotides.
J.Org.Chem., 81, 2016
|
|
3UKC
 
 | | (S)-cEt-BNA decamer structure | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*AP*(1TL)P*AP*CP*GP*C)-3') | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2011-11-09 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structure and nuclease resistance of 2',4'-constrained 2'-O-methoxyethyl (cMOE) and 2'-O-ethyl (cEt) modified DNAs.
Chem.Commun.(Camb.), 48, 2012
|
|
3UKE
 
 | | (S)-cMOE-BNA decamer structure | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*AP*(CSM)P*AP*CP*GP*C)-3') | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2011-11-09 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structure and nuclease resistance of 2',4'-constrained 2'-O-methoxyethyl (cMOE) and 2'-O-ethyl (cEt) modified DNAs.
Chem.Commun.(Camb.), 48, 2012
|
|
3JZM
 
 | | Crystal structure of the phosphorylation-site mutant T432A of the KaiC circadian clock protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein kinase kaiC, MAGNESIUM ION | | Authors: | Pattanayek, R, Egli, M, Pattanayek, S. | | Deposit date: | 2009-09-23 | | Release date: | 2010-03-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of KaiC Circadian Clock Mutant Proteins: A New Phosphorylation Site at T426 and Mechanisms of Kinase, ATPase and Phosphatase.
Plos One, 4, 2009
|
|
3V06
 
 | | Crystal structure of S-6'-Me-3'-fluoro hexitol nucleic acid | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*AP*(F5H)P*AP*CP*GP*C)-3'), STRONTIUM ION | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2011-12-07 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Insights from crystal structures into the opposite effects on RNA affinity caused by the s- and R-6'-methyl backbone modifications of 3'-fluoro hexitol nucleic Acid.
Biochemistry, 51, 2012
|
|
3UKB
 
 | | (R)-cEt-BNA decamer structure | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*AP*(RCE)P*AP*CP*GP*C)-3') | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2011-11-09 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structure and nuclease resistance of 2',4'-constrained 2'-O-methoxyethyl (cMOE) and 2'-O-ethyl (cEt) modified DNAs.
Chem.Commun.(Camb.), 48, 2012
|
|
3K09
 
 | | Crystal structure of the phosphorylation-site mutant S431D of the KaiC circadian clock protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein kinase kaiC, MAGNESIUM ION | | Authors: | Pattanayek, R, Egli, M, Pattanayek, S. | | Deposit date: | 2009-09-24 | | Release date: | 2010-03-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of KaiC Circadian Clock Mutant Proteins: A New Phosphorylation Site at T426 and Mechanisms of Kinase, ATPase and Phosphatase.
Plos One, 4, 2009
|
|
3V07
 
 | | Crystal structure of R-6'-Me-3'-fluoro hexitol nucleic acid | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*AP*(F6H)P*AP*CP*GP*C)-3') | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2011-12-07 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Insights from crystal structures into the opposite effects on RNA affinity caused by the s- and R-6'-methyl backbone modifications of 3'-fluoro hexitol nucleic Acid.
Biochemistry, 51, 2012
|
|
3K0F
 
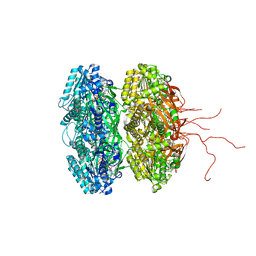 | | Crystal structure of the phosphorylation-site double mutant T426A/T432A of the KaiC circadian clock protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein kinase KaiC, MAGNESIUM ION | | Authors: | Pattanayek, R, Egli, M, Pattanayek, S. | | Deposit date: | 2009-09-24 | | Release date: | 2010-03-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of KaiC Circadian Clock Mutant Proteins: A New Phosphorylation Site at T426 and Mechanisms of Kinase, ATPase and Phosphatase.
Plos One, 4, 2009
|
|
3K0A
 
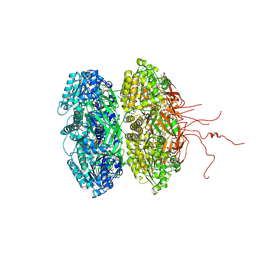 | | Crystal structure of the phosphorylation-site mutant S431A of the KaiC circadian clock protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein kinase KaiC, MAGNESIUM ION | | Authors: | Pattanayek, R, Egli, M, Pattanayek, S. | | Deposit date: | 2009-09-24 | | Release date: | 2010-03-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of KaiC Circadian Clock Mutant Proteins: A New Phosphorylation Site at T426 and Mechanisms of Kinase, ATPase and Phosphatase.
Plos One, 4, 2009
|
|
3K0C
 
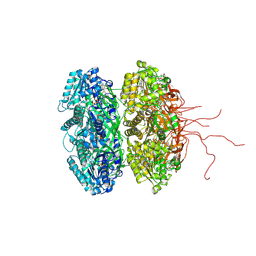 | | Crystal structure of the phosphorylation-site double mutant S431A/T432E of the KaiC circadian clock protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein kinase KaiC, MAGNESIUM ION | | Authors: | Pattanayek, R, Egli, M, Pattanayek, S. | | Deposit date: | 2009-09-24 | | Release date: | 2010-03-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of KaiC Circadian Clock Mutant Proteins: A New Phosphorylation Site at T426 and Mechanisms of Kinase, ATPase and Phosphatase.
Plos One, 4, 2009
|
|
3K0E
 
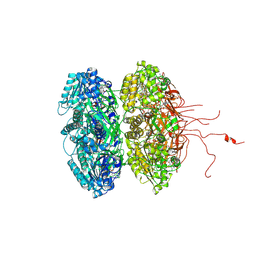 | | Crystal structure of the phosphorylation-site mutant T426N of the KaiC circadian clock protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein kinase KaiC, MAGNESIUM ION | | Authors: | Pattanayek, R, Egli, M, Pattanayek, S. | | Deposit date: | 2009-09-24 | | Release date: | 2010-03-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of KaiC Circadian Clock Mutant Proteins: A New Phosphorylation Site at T426 and Mechanisms of Kinase, ATPase and Phosphatase.
Plos One, 4, 2009
|
|
2QKE
 
 | |
2RF3
 
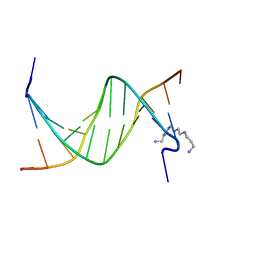 | | Crystal Structure of Tricyclo-DNA: An Unusual Compensatory Change of Two Adjacent Backbone Torsion Angles | | Descriptor: | 5'-d(CGCG(TCY)ATTCGCG)-3', SPERMINE, ZINC ION | | Authors: | Pallan, P.S, Ittig, D, Heroux, A, Wawrzak, Z, Leumann, C.J, Egli, M. | | Deposit date: | 2007-09-27 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of tricyclo-DNA: an unusual compensatory change of two adjacent backbone torsion angles.
Chem.Commun.(Camb.), 2008, 2008
|
|
2R8I
 
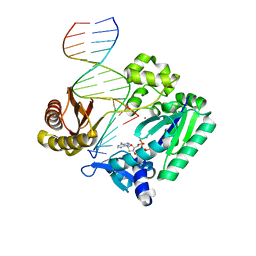 | | Selectivity of Nucleoside Triphosphate Incorporation Opposite 1,N2-Propanodeoxyguanosine (PdG) by the Sulfolobus solfataricus DNA Polymerase Dpo4 Polymerase | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*DGP*DGP*DGP*DGP*DGP*DAP*DAP*DGP*DGP*DAP*DTP*DTP*DC)-3'), ... | | Authors: | Wang, Y, Saleh, S, Marnette, L.J, Egli, M, Stone, M.P. | | Deposit date: | 2007-09-10 | | Release date: | 2008-07-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Insertion of dNTPs opposite the 1,N2-propanodeoxyguanosine adduct by Sulfolobus solfataricus P2 DNA polymerase IV
Biochemistry, 47, 2008
|
|
