7Z7E
 
 | | Crystal structure of p63 DNA binding domain in complex with inhibitory DARPin G4 | | Descriptor: | DARPIN, Isoform 4 of Tumor protein 63, ZINC ION | | Authors: | Strubel, A, Gebel, J, Chaikuad, A, Muenick, P, Doetsch, V. | | Deposit date: | 2022-03-15 | | Release date: | 2022-06-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Designed Ankyrin Repeat Proteins as a tool box for analyzing p63.
Cell Death Differ., 29, 2022
|
|
7Z71
 
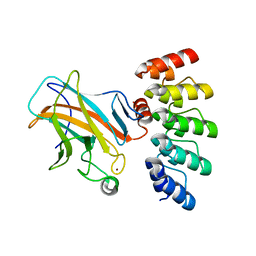 | | Crystal structure of p63 DBD in complex with darpin C14 | | Descriptor: | Darpin C14, Isoform 4 of Tumor protein 63, ZINC ION | | Authors: | Chaikuad, A, Strubel, A, Doetsch, V, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-03-14 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Designed Ankyrin Repeat Proteins as a tool box for analyzing p63.
Cell Death Differ., 29, 2022
|
|
7Z72
 
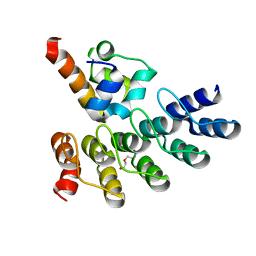 | | Crystal structure of p63 SAM in complex with darpin A5 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Darpin A5, Isoform 9 of Tumor protein 63 | | Authors: | Chaikuad, A, Strubel, A, Doetsch, V, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-03-14 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Designed Ankyrin Repeat Proteins as a tool box for analyzing p63.
Cell Death Differ., 29, 2022
|
|
7Z73
 
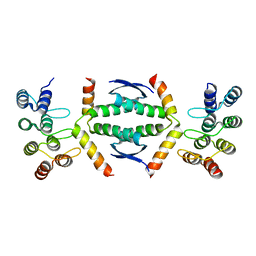 | | Crystal structure of p63 tetramerization domain in complex with darpin 8F1 | | Descriptor: | Darpin 8F1, Isoform 2 of Tumor protein 63 | | Authors: | Chaikuad, A, Strubel, A, Doetsch, V, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-03-14 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Designed Ankyrin Repeat Proteins as a tool box for analyzing p63.
Cell Death Differ., 29, 2022
|
|
7Z7C
 
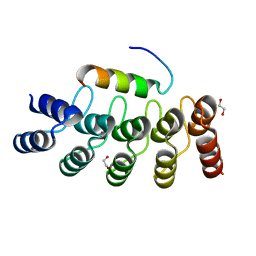 | |
7ZYU
 
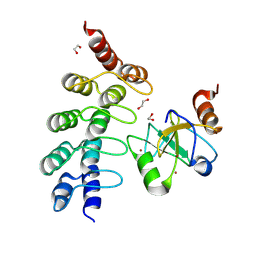 | |
8A19
 
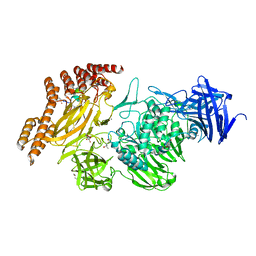 | | Structure of a leucinostatin derivative determined by host lattice display : L1E4V1 construct | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-(2-methoxyethoxy)-11,15-dimethyl-8-oxa-2,11,15,19,21,23-hexazatetracyclo[15.6.1.13,7.020,24]pentacosa-1(23),3(25),4,6,17,20(24),21-heptaen-10-one, ... | | Authors: | Mittl, P.R.E. | | Deposit date: | 2022-06-01 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.358 Å) | | Cite: | Structure of a hydrophobic leucinostatin derivative determined by host lattice display.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
8A1A
 
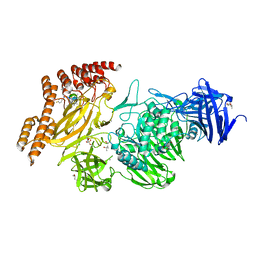 | | Structure of a leucinostatin derivative determined by host lattice display : L1F11V1 construct | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-(2-methoxyethoxy)-11,15-dimethyl-8-oxa-2,11,15,19,21,23-hexazatetracyclo[15.6.1.13,7.020,24]pentacosa-1(23),3(25),4,6,17,20(24),21-heptaen-10-one, ... | | Authors: | Mittl, P.R.E. | | Deposit date: | 2022-06-01 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of a hydrophobic leucinostatin derivative determined by host lattice display.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
8AIW
 
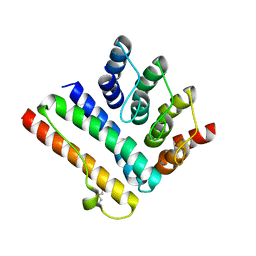 | | Structure of the K5/CagI complex | | Descriptor: | Cag pathogenicity island protein (Cag19), Designed Ankyrin Repeat Protein K5 | | Authors: | Blanc, M, Guerin, J, Terradot, L. | | Deposit date: | 2022-07-27 | | Release date: | 2023-05-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Designed Ankyrin Repeat Proteins provide insights into the structure and function of CagI and are potent inhibitors of CagA translocation by the Helicobacter pylori type IV secretion system.
Plos Pathog., 19, 2023
|
|
8AK1
 
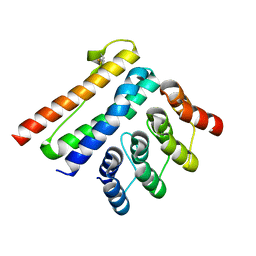 | | Crystal structure of a CagI:K2 complex | | Descriptor: | Cag pathogenicity island protein (Cag19), Designed Ankyrin Repeat Protein K2 | | Authors: | Blanc, M, Guerin, J, Terradot, L. | | Deposit date: | 2022-07-29 | | Release date: | 2023-05-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Designed Ankyrin Repeat Proteins provide insights into the structure and function of CagI and are potent inhibitors of CagA translocation by the Helicobacter pylori type IV secretion system.
Plos Pathog., 19, 2023
|
|
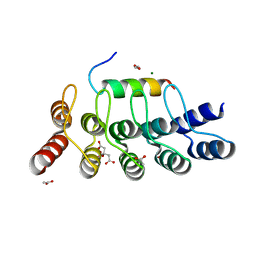
8AED
 
 | |
