3MQ0
 
 | | Crystal Structure of Agobacterium tumefaciens repressor BlcR | | Descriptor: | Transcriptional repressor of the blcABC operon | | Authors: | Chen, L. | | Deposit date: | 2010-04-27 | | Release date: | 2011-03-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.793 Å) | | Cite: | The Agrobacterium tumefaciens Transcription Factor BlcR Is Regulated via Oligomerization.
J.Biol.Chem., 286, 2011
|
|
5ZBG
 
 | |
5YX9
 
 | |
3RJ2
 
 | |
6IMM
 
 | | Cryo-EM structure of an alphavirus, Sindbis virus | | Descriptor: | Assembly protein E3, Octadecane, Spike glycoprotein E1, ... | | Authors: | Zhang, X, Ma, J, Chen, L. | | Deposit date: | 2018-10-23 | | Release date: | 2019-03-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Implication for alphavirus host-cell entry and assembly indicated by a 3.5 angstrom resolution cryo-EM structure.
Nat Commun, 9, 2018
|
|
7D4P
 
 | | Structure of human TRPC5 in complex with clemizole | | Descriptor: | (2S)-2-(hexadecanoyloxy)-3-hydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 1-[(4-chlorophenyl)methyl]-2-(pyrrolidin-1-ylmethyl)benzimidazole, ... | | Authors: | Chen, L, Song, K, Wei, M, Guo, W. | | Deposit date: | 2020-09-24 | | Release date: | 2021-03-31 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis for human TRPC5 channel inhibition by two distinct inhibitors.
Elife, 10, 2021
|
|
7D4Q
 
 | | Structure of human TRPC5 in complex with HC-070 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 8-(3-chloranylphenoxy)-7-[(4-chlorophenyl)methyl]-3-methyl-1-(3-oxidanylpropyl)purine-2,6-dione, CALCIUM ION, ... | | Authors: | Chen, L, Song, K, Wei, M, Guo, W. | | Deposit date: | 2020-09-24 | | Release date: | 2021-03-31 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Structural basis for human TRPC5 channel inhibition by two distinct inhibitors.
Elife, 10, 2021
|
|
7E4T
 
 | | Human TRPC5 apo state structure at 3 angstrom | | Descriptor: | (2S)-2-(hexadecanoyloxy)-3-hydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CALCIUM ION, ... | | Authors: | Chen, L, Song, K, Wei, M, Guo, W. | | Deposit date: | 2021-02-15 | | Release date: | 2021-03-31 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for human TRPC5 channel inhibition by two distinct inhibitors.
Elife, 10, 2021
|
|
7E28
 
 | |
7E24
 
 | |
8K5G
 
 | |
8K5H
 
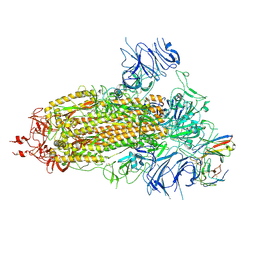 | | Structure of the SARS-CoV-2 BA.1 spike with UT28-RD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Chen, L, Kita, S, Anraku, Y, Maenaka, K. | | Deposit date: | 2023-07-21 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | In silico-designed UT28K with two amino acid substitutions is capable of neutralization of resistant SARS-CoV2 Omicrons BA.1 valiant.
To Be Published
|
|
6L48
 
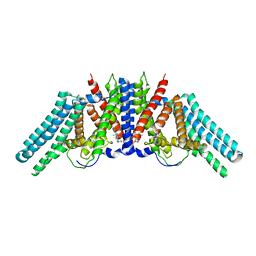 | |
6L47
 
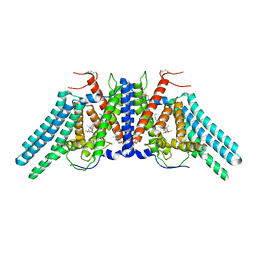 | | Structure of the human sterol O-acyltransferase 1 in complex with CI-976 | | Descriptor: | 2,2-dimethyl-N-(2,4,6-trimethoxyphenyl)dodecanamide, CHOLESTEROL, Sterol O-acyltransferase 1 | | Authors: | Chen, L, Guan, C, Niu, Y. | | Deposit date: | 2019-10-16 | | Release date: | 2020-04-29 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into the inhibition mechanism of human sterol O-acyltransferase 1 by a competitive inhibitor.
Nat Commun, 11, 2020
|
|
7Y4S
 
 | | Structure of human MG53 homo-dimer | | Descriptor: | Tripartite motif-containing protein 72 | | Authors: | Chen, L, Niu, Y. | | Deposit date: | 2022-06-16 | | Release date: | 2022-09-21 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of human MG53 homodimer.
Biochem.J., 479, 2022
|
|
7L7S
 
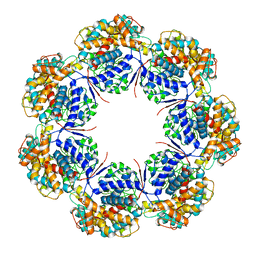 | | Human mitochondrial chaperonin mHsp60 | | Descriptor: | 60 kDa heat shock protein, mitochondrial | | Authors: | Chen, L, Wang, J.C.Y. | | Deposit date: | 2020-12-30 | | Release date: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for the structural dynamics of human mitochondrial chaperonin mHsp60.
Sci Rep, 11, 2021
|
|
5YKF
 
 | |
5YKG
 
 | |
5YKE
 
 | |
5YWC
 
 | |
5YW9
 
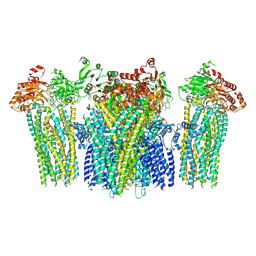 | |
5YWB
 
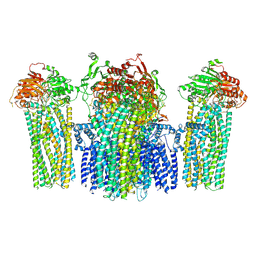 | |
5YW8
 
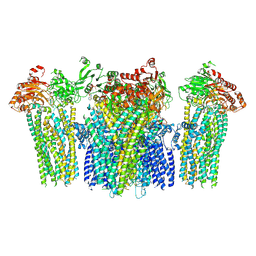 | |
5Z1F
 
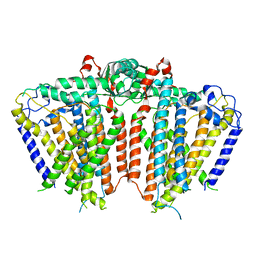 | |
5YWD
 
 | |
